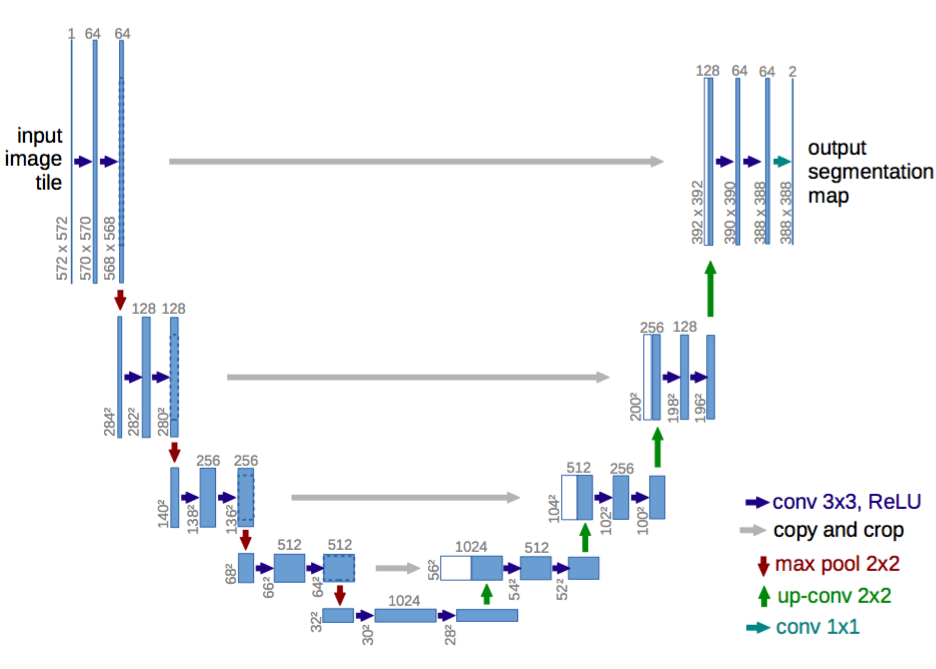
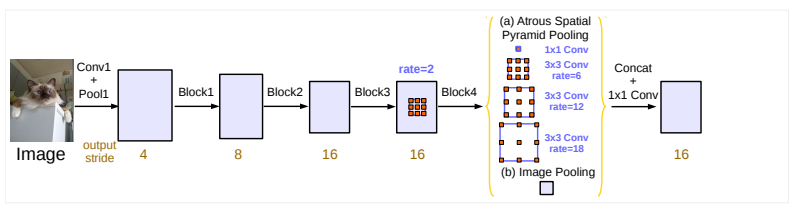
This project includes SegNet, UNet, and DeepLabV3 for Semantic Segmentation of nuclei in digital histology images.
"Semantic segmentation, or image segmentation, is the task of clustering parts of an image together which belong to the same object class. It is a form of pixel-level prediction because each pixel in an image is classified according to a category. Some example benchmarks for this task are Cityscapes, PASCAL VOC and ADE20K. Models are usually evaluated with the Mean Intersection-Over-Union (Mean IoU) and Pixel Accuracy metrics." -- PapersWithCode.
MoNuSeg dataset (available here) contains multi organ tissue images with the ground truth segmentation masks for nuclei. The dataset can also be downloaded from this Google Drive link.
Following is a sample training Tissue image with its correcsponding ground truth segmentation mask.
The dataset contains 30 training, 14 testing samples.
Tissue images shapes: 1000x1000x3
Ground truth segmentation masks shapes: 1000x1000x3
- The training data is split into training and validation sets with 75:25 ratio.
- Ground truth segmentation masks are reshaped such that only one channal of each is kept.
- Both the training and validation images and their correcsponding ground truth segmentation masks are split into 256x256 subimages using sliding-window approch with around 20% overlap ratio using a custom data spliter.
- Training images and their correcsponding ground truth segmentation masks are padded with zeroes such that we get 1024x1024 size images. This is done to avoid model crash or to avoid getting output shape lesser than that of input.
- After getting predictions on these padded test images, the paddings are removed from predictions and test data to get performance measures.
Following three augmentations are applied on the training and validation images and their correcsponding ground truth segmentation masks using a custom data augmenter:
- Rotations of angles 90, 180, 270 degrees.
- Horizontal flips
- Vertical flips
These augmentations were applied on 10% of the training and 20% of the validation data.
- python version 3.x
- tensorflow version 1.x
.
├── Colab Notebooks # Interactive notebooks for training, testing, and predictions
├── Configs # Configuration files for respective models
├── Images # Screenshots or images needed for better presentation of README.md file
├── Models # Complete implementations of models of the project
│ ├── DeepLabV3.py # DeepLabV3 standard model
│ ├── DeepLabV3Plus.py # DeepLabV3+ standard model
│ ├── SegNet.py # SegNet standard model
│ ├── SegNet_ResNet.py # SegNet model with ResNet backbones
│ ├── UNet.py # UNet standard model
│ └── UNet_ResNet.py # UNet model with ResNet backbones
├── Training Plots # Training and validation performance graphs for loss, accuracy, and f1 scores
├── Utils # Files that include custom functionalities needed for this project
├── README.md # A complete overview of this directory
└── train.py # Functions to train a model with simple or augmented data
Go to the colab notebooks in the Colab Notebooks directory for each model to view the detailed model summary.
F1 Score is defined as the harmonic mean of precision and recall as where
and
. This is equivalent to Dice score coefficient which is defined as
.
| Model | Accuracy | Precision | Recall | F1 Score (Dice Score) |
|---|---|---|---|---|
| SegNet | 0.9138 | 0.79 | 0.77 | 0.7799 |
| UNet | 0.9048 | 0.72 | 0.86 | 0.7802 |
| DeepLabV3 | 0.8606 | 0.61 | 0.80 | 0.6943 |
| SegNet_ResNet | 0.9042 | 0.74 | 0.81 | 0.7686 |
| UNet_ResNet | 0.9154 | 0.76 | 0.83 | 0.7942 |
| DeepLabV3Plus | 0.8988 | 0.72 | 0.80 | 0.7576 |
Following is the test tissue image with its ground truth segmentation mask that I show the qualitative results of my models on.
Use the colab notebooks in the Colab Notebooks directory for training, testing, and predictions on different models.
- SegNet_basic.model
- UNet_basic.model
- DeepLabV3_basic.model
- SegNet_resnet.model
- UNet_resnet.model
- DeepLabV3Plus.model
Either use the colab notebooks in the Colab Notebooks directory for predictions on respective models, or follow the following steps using your console.
git clone https://github.com/zeeshanalipanhwar/Semantic-Segmentation-Keras
mv Semantic-Segmentation-Keras Semantic_Segmentation_Keras
# import all the models and their respective configuration files
from Semantic_Segmentation_Keras.Models import SegNet, UNet, DeepLabV3, SegNet_ResNet, UNet_ResNet, DeepLabV3Plus
from Semantic_Segmentation_Keras.Configs import SegNet_Configs, UNet_Configs, DeepLabV3_Configs# create a model of your choice among the above availabe models
model = SegNet.SegNet(depth=SegNet_Configs.DEPTH).SegNet(input_shape=(None,None, 3))
#model = UNet.UNet(depth=UNet_Configs.DEPTH).UNet(input_shape=(None, None, 3))
#model = DeepLabV3.DeepLabV3(depth=DeepLabV3_Configs.DEPTH).DeepLabV3(input_shape=(None, None, 3))
#model = SegNet_ResNet.SegNet_ResNet(depth=SegNet_Configs.DEPTH).SegNet_ResNet(input_shape=(None,None, 3))
#model = UNet_ResNet.UNet_ResNet(depth=UNet_Configs.DEPTH).UNet_ResNet(input_shape=(None, None, 3))
#model = DeepLabV3Plus.DeepLabV3Plus(depth=DeepLabV3_Configs.DEPTH).DeepLabV3Plus(input_shape=(None, None, 3))# optionally view the created model summary
model.summary()model.load_weights("SegNet_basic.model")
#model.load_weights("UNet_basic.model")
#model.load_weights("DeepLabV3_basic.model")
#model.load_weights("SegNet_resnet.model")
#model.load_weights("UNet_resnet.model")
#model.load_weights("DeepLabV3Plus.model")from Semantic_Segmentation_Keras.Utils import display
import numpy as np
import cv2
print_statements = False # do you need to see the print results blow?
# load a sample image
image_path = "drive/My Drive/sample_tissue_image.tif"
sample_image = cv2.cvtColor(cv2.imread(image_path), cv2.COLOR_BGR2RGB)
sample_image = np.array(sample_image, dtype="float") / 255.0
sample_image = np.expand_dims(sample_image, axis=0)
if print_statements: print ("sample_image shape:", sample_image.shape)
# in order to avoid a crash of model, make sure the image spatial dimentions are a multiple of 16
# the multiple factor 16 represented the ratio to which the actual image is reduced to by a model
padx, pady = 0, 0 # number zeros to add in x and y respectively
origional_sample_image_shape = sample_image.shape
if print_statements: print ("origional_sample_image_shape", origional_sample_image_shape)
if sample_image.shape[1]//16 != sample_image.shape[1]/16:
padx = int(2**round(np.log2(sample_image.shape[1]))-sample_image.shape[1])//2
if sample_image.shape[2]//16 != sample_image.shape[2]/16:
pady = int(2**round(np.log2(sample_image.shape[2]))-sample_image.shape[2])//2
if print_statements: print ("padx={}, pady={}".format(padx, pady))
sample_image_padded = np.zeros((1, sample_image.shape[1]+2*padx,
sample_image.shape[2]+2*pady, 3))
if print_statements: print ("sample_image_padded shape", sample_image_padded.shape)
sample_image_padded[:, padx:padx+sample_image.shape[1],
pady:pady+sample_image.shape[2], :] = sample_image
sample_image = sample_image_padded
if print_statements: print ("sample_image shape:", sample_image.shape)
# make prediction for a sample on the network
prediction = model.predict(sample_image)
prediction = prediction.round(0)
if print_statements: print ("prediction shape:", prediction.shape)
# discard the predictions for the padded portion of sample_image
prediction = prediction[:, padx:padx+origional_sample_image_shape[1],
pady:pady+origional_sample_image_shape[2], :]
if print_statements: print ("prediction shape:", prediction.shape)
# remove the padded zeros from sample_image
sample_image = sample_image[:, padx:padx+origional_sample_image_shape[1],
pady:pady+origional_sample_image_shape[2], :]
if print_statements: print ("sample_image shape:", sample_image.shape)
# display the sample image along with its predicted mask
display.display_masked(sample_image[0], prediction[0], "Tissue Image", "Predicted Mask")This project is licensed under the terms of the MIT License.
The ./Utils/custom_layers.py contains updated classes from ykamikawa/tf-keras-SegNet/layers.py file.
This project structure followed guidlines from DongjunLee/hb-base repository.
The ./.github/CONTRIBUTING.md was adapted from a basic template for contributing guidelines.
The ./.github/PULL_REQUEST_TEMPLATE.md is taken from TalAter/open-source-templates.
Maintainer Zeeshan Ali ([email protected])