-
Notifications
You must be signed in to change notification settings - Fork 4
1.2. Output Contents (FastDTLmapper)
In this page, explain output contents of following 5 files in 03_aggregate_map_result.
.
├── 00_user_data/
├── 01_orthofinder/
├── 02_dtl_reconciliation/
├── 03_aggregate_map_result/ -- Genome-wide DTL reconciliation aggregated and mapped results
│ ├── all_dtl_map.nwk -- Genome-wide DTL event mapped tree file
│ ├── all_gain_loss_map.nwk -- Genome-wide Gain-Loss event mapped tree file
│ ├── all_og_node_event.tsv -- All OG DTL event record file
│ ├── all_transfer_gene_count.tsv -- All transfer gene count file
│ └── all_transfer_gene_list.tsv -- All transfer gene list file
└── log/
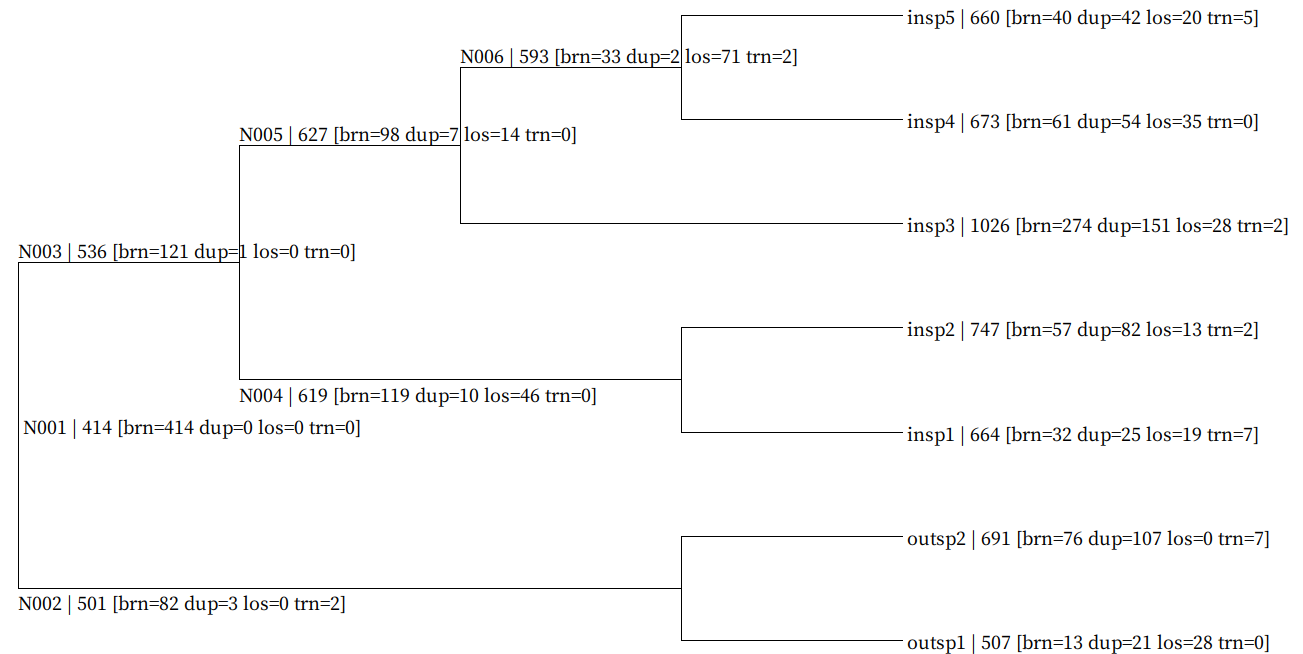
Genome-wide DTL event mapped tree file.
Each node DTL map data is described in following format.
NodeID | GeneNum [brn=BornNum dup=DuplicationNum los=LossNum trn=TransferNum]
Example of all_dtl_map.nwk:
((outsp1 | 507 [brn=13 dup=21 los=28 trn=0]:0.107239,outsp2 | 691 [brn=76 dup=107 los=0 trn=7]:0.107239)N002 | 501 [brn=82 dup=3 los=0 trn=2]:0.306859,((insp1 | 664 [brn=32 dup=25 los=19 trn=7]:0.027931,insp2 | 747 [brn=57 dup=82 los=13 trn=2]:0.027931)N004 | 619 [brn=119 dup=10 los=46 trn=0]:0.324618,(insp3 | 1026 [brn=274 dup=151 los=28 trn=2]:0.243408,(insp4 | 673 [brn=61 dup=54 los=35 trn=0]:0.113100,insp5 | 660 [brn=40 dup=42 los=20 trn=5]:0.113100)N006 | 593 [brn=33 dup=2 los=71 trn=2]:0.130308)N005 | 627 [brn=98 dup=7 los=14 trn=0]:0.109141)N003 | 536 [brn=121 dup=1 los=0 trn=0]:0.061550)N001 | 414 [brn=414 dup=0 los=0 trn=0];
Use SeaView to visualize it as shown below.
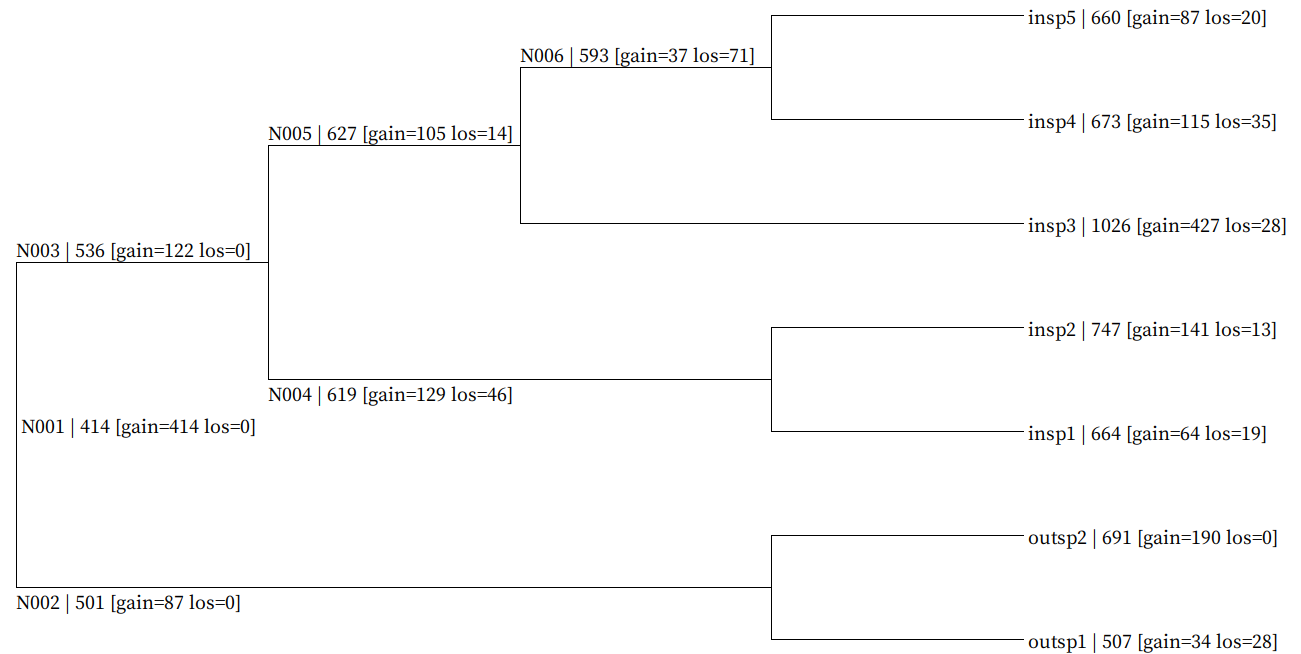
Genome-wide Gain-Loss event mapped tree file.
Each node gain/loss map data is described in following format.
NodeID | GeneNum [gain=GainNum los=LossNum]
Example of all_gain_loss_map.nwk:
((outsp1 | 507 [gain=34 los=28]:0.107239,outsp2 | 691 [gain=190 los=0]:0.107239)N002 | 501 [gain=87 los=0]:0.306859,((insp1 | 664 [gain=64 los=19]:0.027931,insp2 | 747 [gain=141 los=13]:0.027931)N004 | 619 [gain=129 los=46]:0.324618,(insp3 | 1026 [gain=427 los=28]:0.243408,(insp4 | 673 [gain=115 los=35]:0.113100,insp5 | 660 [gain=87 los=20]:0.113100)N006 | 593 [gain=37 los=71]:0.130308)N005 | 627 [gain=105 los=14]:0.109141)N003 | 536 [gain=122 los=0]:0.061550)N001 | 414 [gain=414 los=0];
Use SeaView to visualize it as shown below.
All OG(Ortholog Group) DTL event record tsv format file (See example data here).
| Columns | Contents |
|---|---|
| OG_ID | OG(Ortholog Group) ID generated by OrthoFinder |
| NODE_ID | Species tree node id |
| GENE_NUM | OG-Node gene number |
| GAIN_NUM | OG-Node gain number (Gain=Brn+Dup+Trn) |
| BRN_NUM | OG-Node born number |
| DUP_NUM | OG-Node duplication number |
| TRN_NUM | OG-Node transfer number |
| LOS_NUM | OG-Node loss number |
| TRN_DETAIL | Transer path list (e.g. 'N023 -> N012,N011 -> N008') |
All transfer gene count tsv format file (See example data here).
Gene transfer information is extracted from AnGST result file.
| Columns | Contents |
|---|---|
| TRANSFER_PATH | Transfer path (e.g. 'N023 -> N012') |
| TRANSFER_DERIVED_GENE_NUM | Transfer derived gene number in TRANSFER_PATH |
| TRANSFER_DERIVED_GENE_ID_LIST | Transfer derived gene id list in TRANSFER_PATH |
All transfer gene list tsv format file (See example data here).
Gene transfer information is extracted from AnGST result file.
| Columns | Contents |
|---|---|
| OG_ID | OG(Ortholog Group) ID |
| TRANSFER_DERIVED_GENE_ID | Transfer derived gene id |
| TRANSFER_PATH | Transfer path (e.g. 'N023 -> N012') |
- FastDTLmapper
- FastDTLgoea (subtool)
- plot_gain_loss_map (subtool)