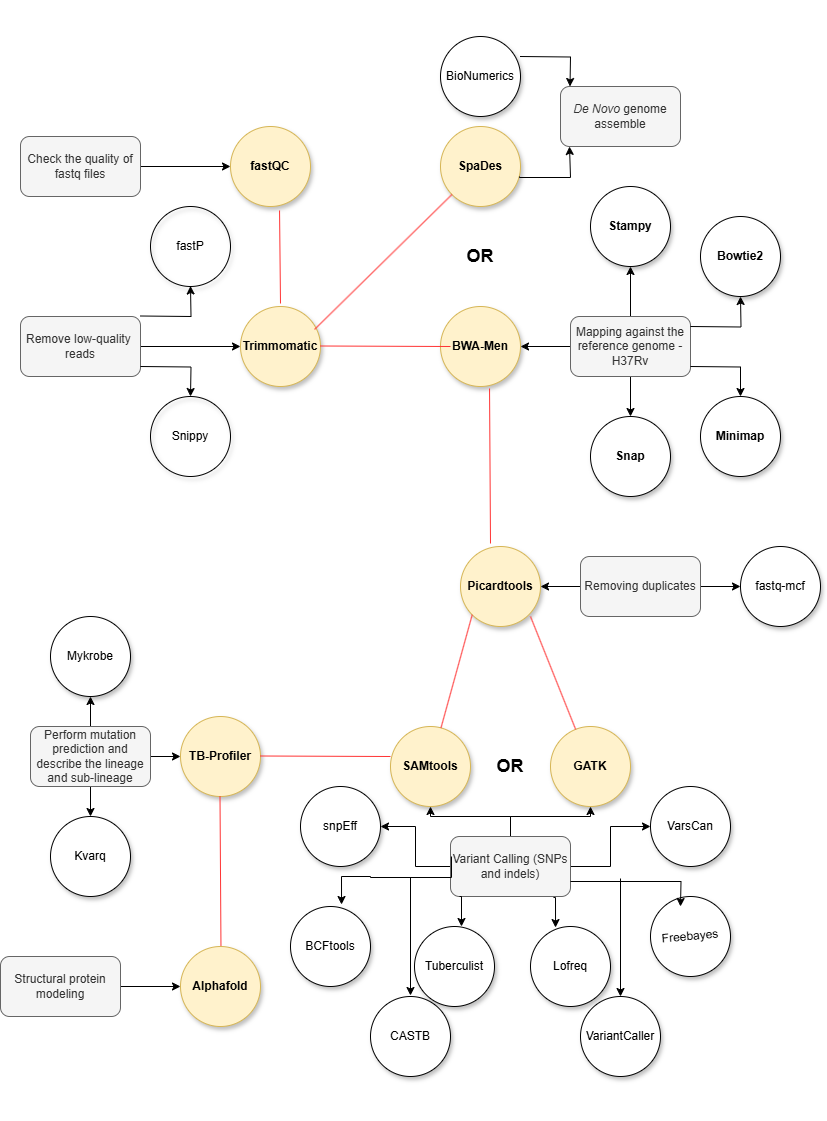
Click to reveal the text
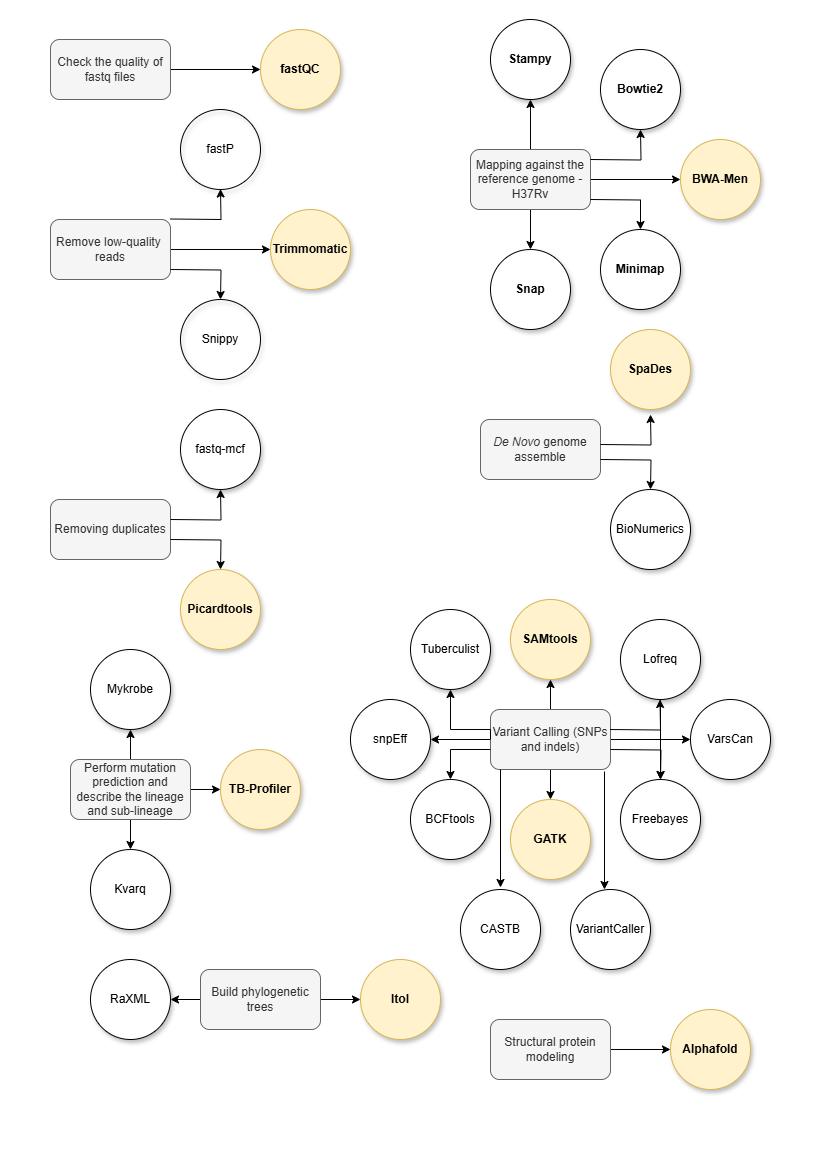
- Check the quality of FASTQ files
- Remove low-quality reads
- Remove duplicates
- Map against the H37Rv genome or perform De Novo assembly
- Variant calling (SNPs and indels)
- Classify resistance-related mutations
- Compare with phenotypic data for validation
Click to reveal the text
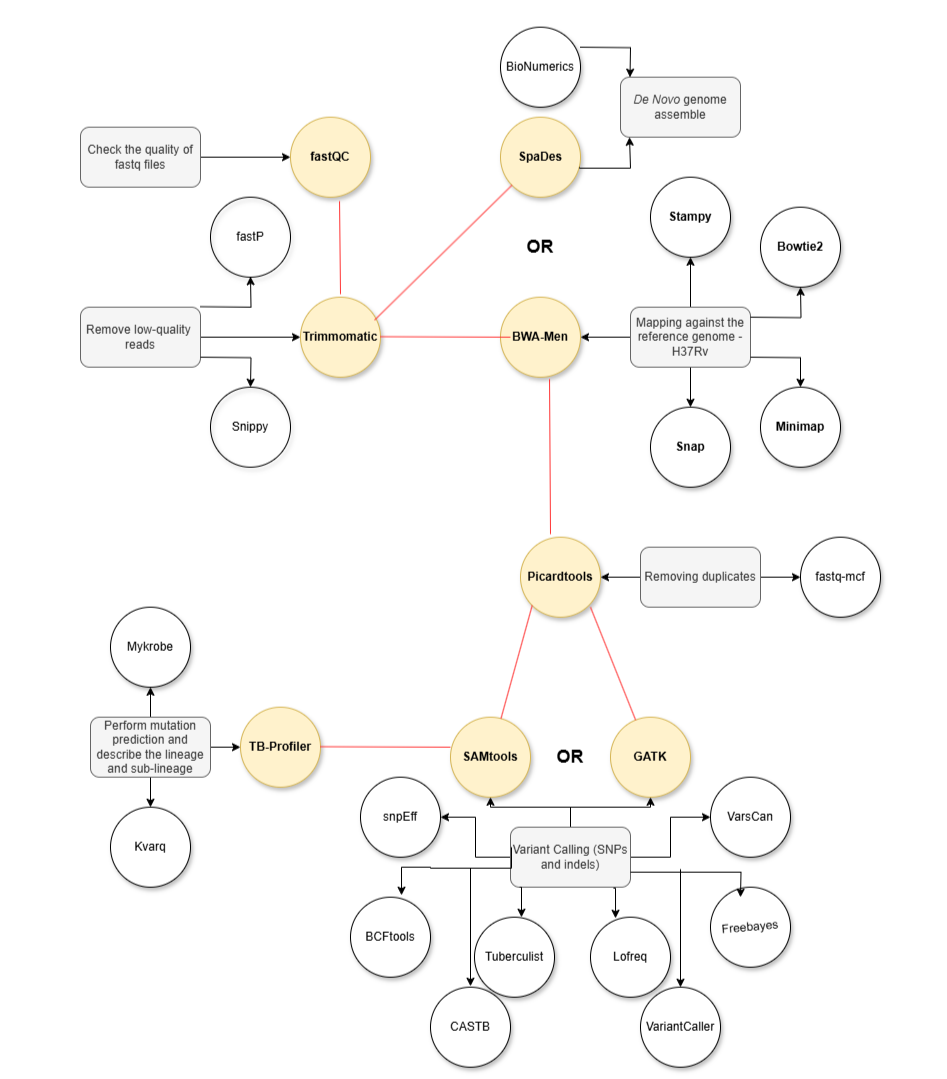
- Check the quality of FASTQ files
- Remove low-quality reads
- Remove duplicates
- Map against the H37Rv genome or perform De Novo assembly
- Variant calling (SNPs and indels)
- Identify variants not listed in databases
- Associate with specific genes
Click to reveal the text
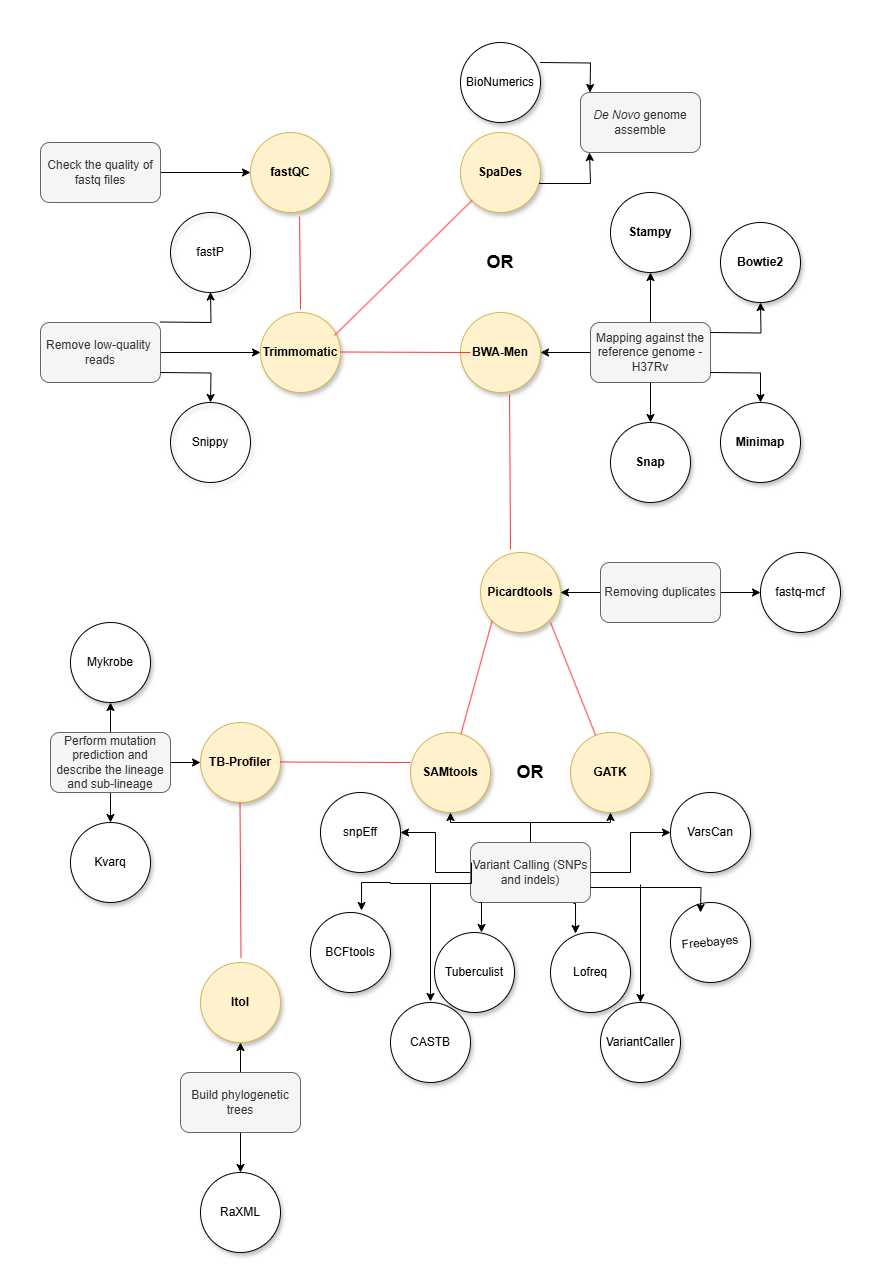
- Check the quality of FASTQ files
- Remove low-quality reads
- Remove duplicates
- Map against the H37Rv genome or perform De Novo assembly
- Phylogenetic analysis
- Build phylogenetic trees
- Genome comparison
- Analysis of conserved and convergent regions
Click to reveal the text
- Check the quality of FASTQ files
- Remove low-quality reads
- Remove duplicates
- Map against the H37Rv genome or perform De Novo assembly
- Identification of specific genes
- Structural protein modeling
- Virtual screening
- Simulate interaction with drugs
- Interactive Workflow: Click on the images to navigate to the corresponding section.
- Back to Top Links: Each section includes a link to return to the top of the page.
- Clear Steps: Each step is outlined in a simple, easy-to-follow format.
For any questions, please contact me by email: [email protected]