A comprehensive pipeline for panoptic segmentation and classification of nuclei in histopathology images using Pathopix-GANs, Segment Anything (SAM), Diffusion Models, and YOLOSeg.
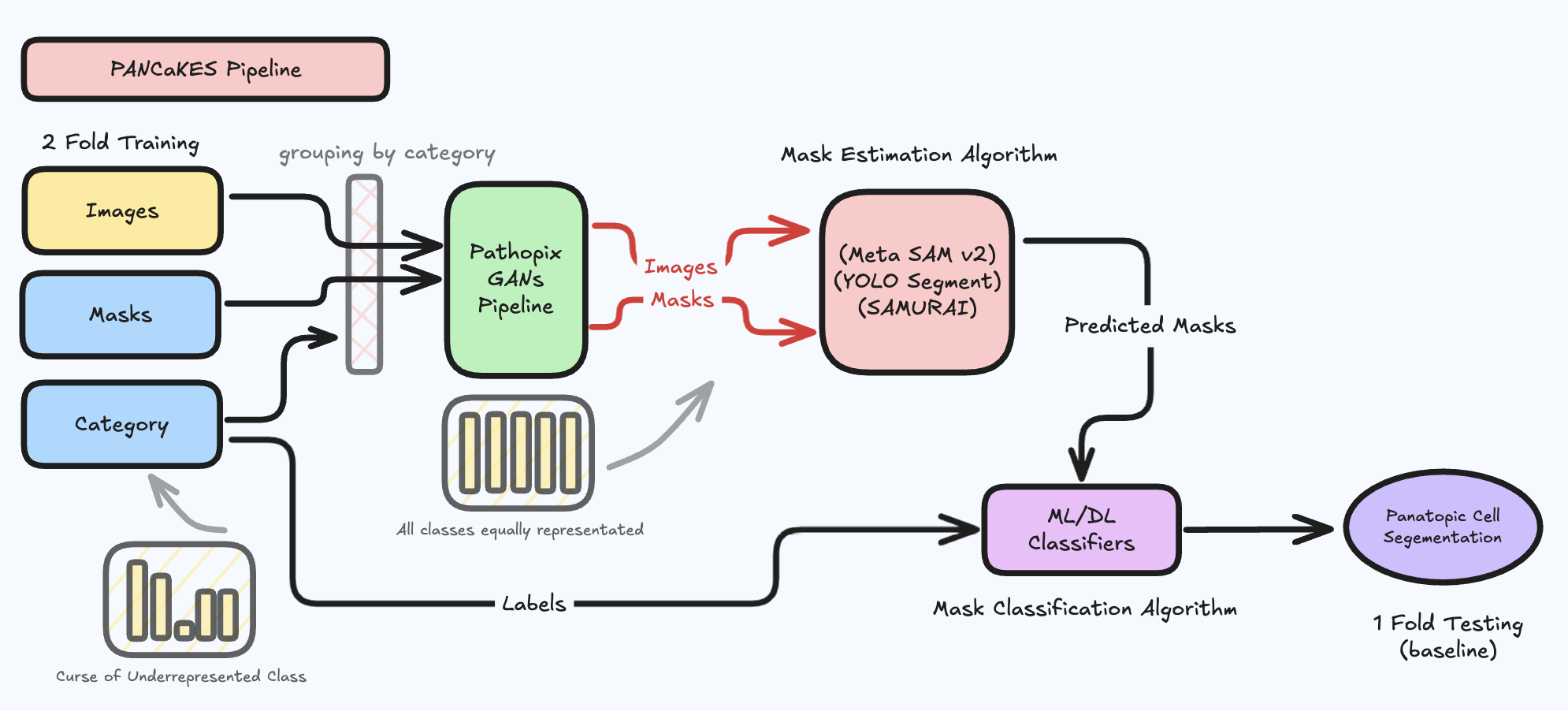
PANCaKES explores a multi-model strategy to tackle the challenging task of panoptic cell segmentation, leveraging:
- Adversarial training using Pathopix-GANs
- Foundation models like SAM and Segment-Transformer
- End-to-end YOLOSeg fine-tuning for instance segmentation
- Hybrid data augmentation and GAN-generated synthetic samples
The pipeline spans data preprocessing, exploratory analysis, training, evaluation, and inference across different model strategies.
The high-level architecture of the full segmentation-classification pipeline is shown below:
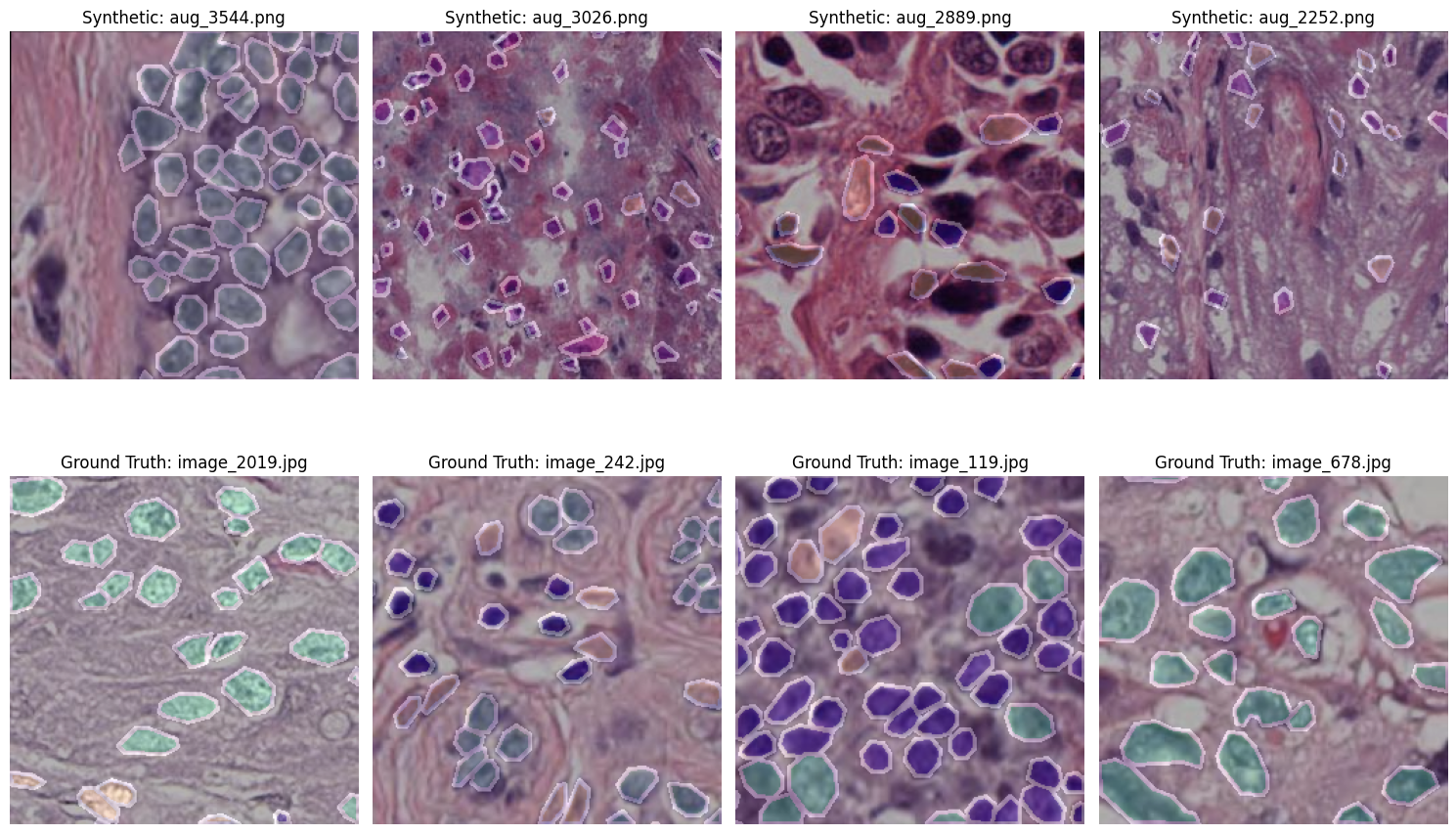
Below are sample outputs at different epochs during GAN training. These reflect the model's generative progress from noise to realistic nuclei masks/images.
| Epoch | Output |
|---|---|
| Epoch 100 | 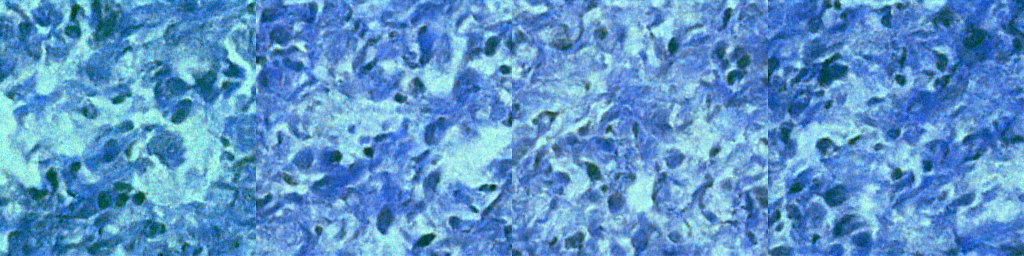 |
| Epoch 200 | 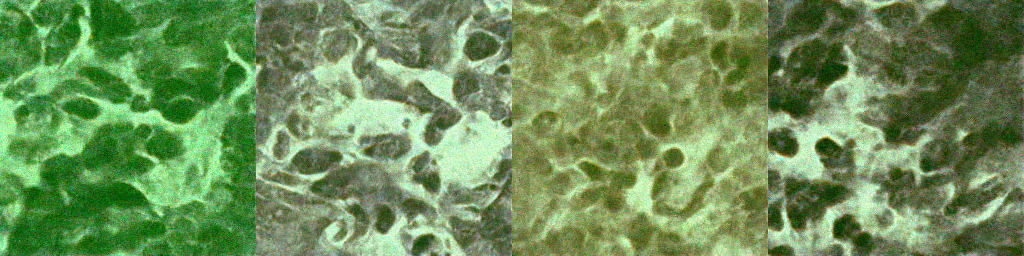 |
| Epoch 300 |  |
These are samples generated using the trained GAN model for augmentation:
- End-to-end segmentation & classification
- GAN-based synthetic data augmentation
- Finetuning of SAM, Segment-Transformer, YOLOSeg
- Extensive visual analytics and metrics
- COCO ↔ YOLO format conversion scripts
- Organized cookbooks for reproducibility
├── cookbooks/ # All model training + EDA notebooks
├── data/ # Dataset folders with folds and masks
├── docs/ # Project proposal and deliverables
├── imgs/ # Visual outputs and result images
├── results/ # Metrics and logs
├── scripts/ # Format conversion & data generation
├── README.md
└── LICENSE| Notebook | Description |
|---|---|
[0] Data Loader.ipynb |
Data exploration & loader setup |
[1] EDA.ipynb |
Visual EDA and class distribution |
[2] Data Transformation & Upload.ipynb |
Preprocessing and upload routines |
[3] PP-GANs Pipeline (Failed).ipynb |
Attempted GANs-based augmentation |
[4] GANs-Training.ipynb |
Training GANs for synthetic nuclei |
[5] COCO-Format-Conversion.ipynb |
JSON annotation transformation |
[6] GANs-Augmentation.ipynb |
Applying trained GANs to dataset |
[7] Finetune-SAMv2.ipynb |
Fine-tuning SAM |
[8] Finetune-YoloSeg.ipynb |
Fine-tuning YOLOSeg |
[9] YoloSeg-Inference.ipynb |
Inference on test data using YOLOSeg |
[10] Finetune-SAM+SegTf.ipynb |
SAM + Segment Transformer |
[11] SAM+SegTf-Inference.ipynb |
Inference with SAM + SegTf |
| Sample | Output |
|---|---|
| YOLOSeg - Sample 1 | 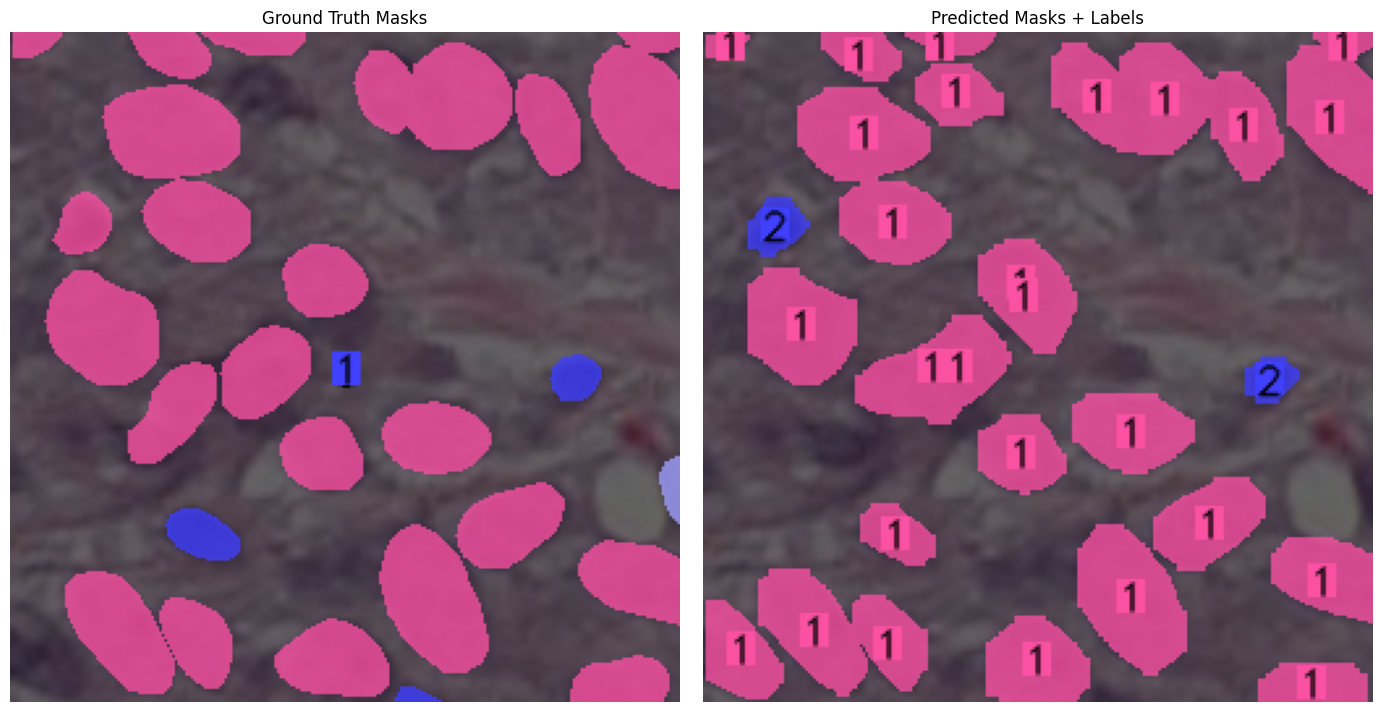 |
| YOLOSeg - Sample 2 | 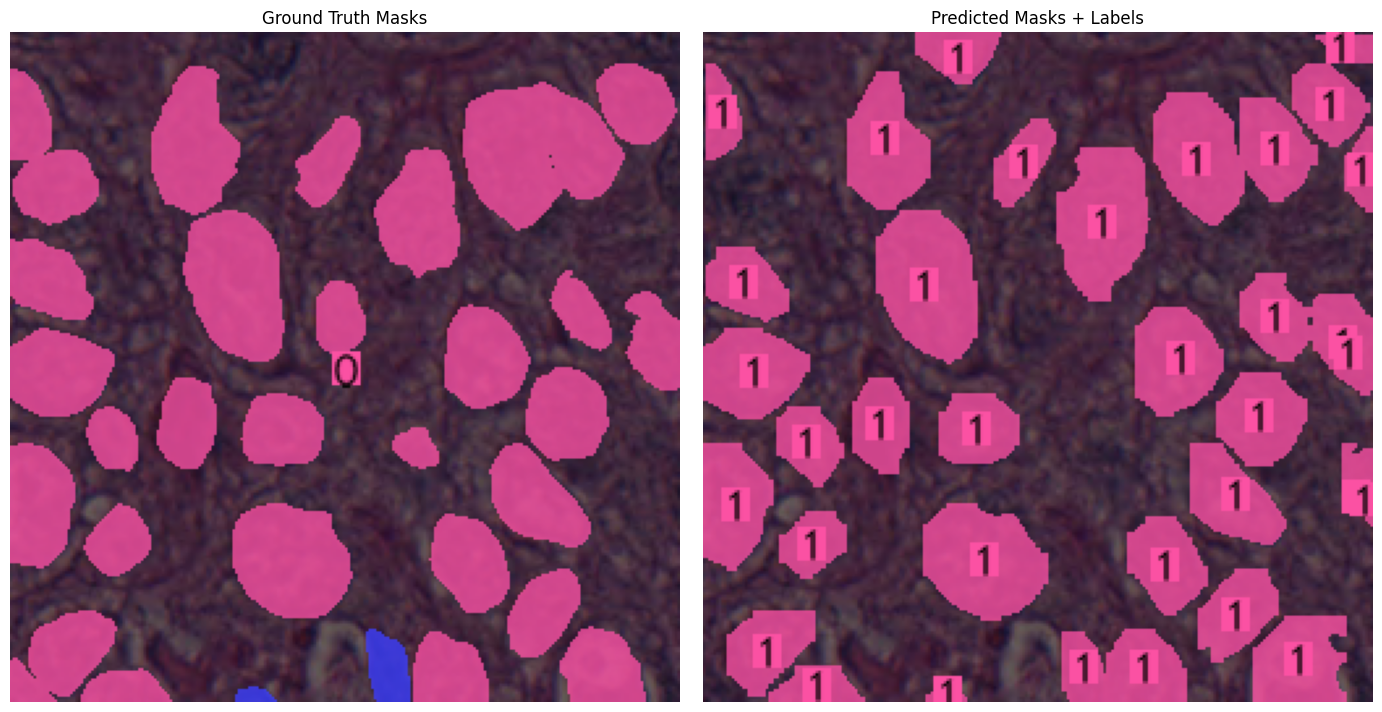 |
- Performance results are logged in
results/metrics.xlsx - Includes segmentation metrics: mIoU, Dice Coefficient, Precision, Recall
- Also compares across folds (fold_1, fold_2, fold_3)
Install necessary dependencies using:
pip install -r requirements.txtOr manually include major packages:
torch,torchvision,transformers,opencv-pythonmatplotlib,numpy,pandas,tqdm,pycocotools
Refer to the following PDFs in the docs/ directory:
proposal.pdf: Project background and goalsdeliverable-i.pdf: Phase I progress and resultsdeliverable-ii.pdf: Final results and summary
Each fold contains sample images with corresponding mask files.
data/fold_1/sample_0/
├── image.png
└── mask/
├── mask_0_0.png
└── mask_0_1.png
Utility scripts for data format conversion and generation:
| Script | Purpose |
|---|---|
scripts/coco2yolo.py |
Convert COCO format annotations to YOLO format |
scripts/generate.py |
Custom data generation/augmentation pipeline |
We welcome PRs, issue reports, and feedback! Feel free to fork, star 🌟, and share.
Licensed under the MIT License.