| Sample Plot 1 | +Sample Plot 2 | +
|---|---|
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+  +
+ |
+
+ 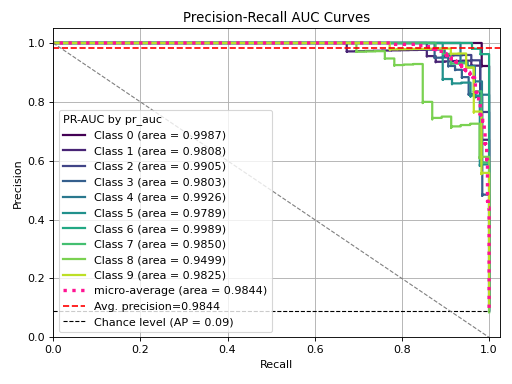 +
+
-Finally, compare and [view the non-scikit-plot way of plotting the multi-class ROC curve](http://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.html). Which one would you rather do?
+Pretty.
## Maximum flexibility. Compatibility with non-scikit-learn objects.
-Although Scikit-plot is loosely based around the scikit-learn interface, you don't actually need Scikit-learn objects to use the available functions. As long as you provide the functions what they're asking for, they'll happily draw the plots for you.
-
-Here's a quick example to generate the precision-recall curves of a Keras classifier on a sample dataset.
-
-```python
-# Import what's needed for the Functions API
-import matplotlib.pyplot as plt
-import scikitplot as skplt
-
-# This is a Keras classifier. We'll generate probabilities on the test set.
-keras_clf.fit(X_train, y_train, batch_size=64, nb_epoch=10, verbose=2)
-probas = keras_clf.predict_proba(X_test, batch_size=64)
-
-# Now plot.
-skplt.metrics.plot_precision_recall_curve(y_test, probas)
-plt.show()
-```
-
-
-You can see clearly here that `skplt.metrics.plot_precision_recall_curve` needs only the ground truth y-values and the predicted probabilities to generate the plot. This lets you use *anything* you want as the classifier, from Keras NNs to NLTK Naive Bayes to that groundbreaking classifier algorithm you just wrote.
+Although Scikit-plot is loosely based around the scikit-learn interface, you don't actually need scikit-learn objects to use the available functions.
+As long as you provide the functions what they're asking for, they'll happily draw the plots for you.
The possibilities are endless.
-## Installation
+## User Installation
-Installation is simple! First, make sure you have the dependencies [Scikit-learn](http://scikit-learn.org) and [Matplotlib](http://matplotlib.org/) installed.
+1. **Install Scikit-plots**:
+ - Use pip to install Scikit-plots:
-Then just run:
-```bash
-pip install scikit-plot
-```
+ ```bash
+ pip install scikit-plots
+ ```
-Or if you want the latest development version, clone this repo and run
-```bash
-python setup.py install
-```
-at the root folder.
+## Release Notes
-If using conda, you can install Scikit-plot by running:
-```bash
-conda install -c conda-forge scikit-plot
-```
+See the [changelog](https://scikit-plots.github.io/stable/whats_new/whats_new.html)
+for a history of notable changes to scikit-plots.
## Documentation and Examples
Explore the full features of Scikit-plot.
-You can find detailed documentation [here](http://scikit-plot.readthedocs.io).
-
-Examples are found in the [examples folder of this repo](examples/).
-
-## Contributing to Scikit-plot
-
-Reporting a bug? Suggesting a feature? Want to add your own plot to the library? Visit our [contributor guidelines](CONTRIBUTING.md).
-
-## Citing Scikit-plot
-
-Are you using Scikit-plot in an academic paper? You should be! Reviewers love eye candy.
-
-If so, please consider citing Scikit-plot with DOI [](https://doi.org/10.5281/zenodo.293191)
-
-#### APA
-
-> Reiichiro Nakano. (2018). reiinakano/scikit-plot: 0.3.7 [Data set]. Zenodo. http://doi.org/10.5281/zenodo.293191
-
-#### IEEE
+## Contributing to scikit-plots
-> [1]Reiichiro Nakano, “reiinakano/scikit-plot: 0.3.7”. Zenodo, 19-Feb-2017.
+Reporting a bug? Suggesting a feature? Want to add your own plot to the library? Visit our.
-#### ACM
+## Citing scikit-plots
-> [1]Reiichiro Nakano 2018. reiinakano/scikit-plot: 0.3.7. Zenodo.
+1. scikit-plots, “scikit-plots: vlatest”. Zenodo, Aug. 23, 2024.
+ DOI: [10.5281/zenodo.13367000](https://doi.org/10.5281/zenodo.13367000).
-Happy plotting!
+2. scikit-plots, “scikit-plots: v0.3.8dev0”. Zenodo, Aug. 23, 2024.
+ DOI: [10.5281/zenodo.13367001](https://doi.org/10.5281/zenodo.13367001).
\ No newline at end of file
diff --git a/SECURITY.md b/SECURITY.md
new file mode 100644
index 0000000..d46888a
--- /dev/null
+++ b/SECURITY.md
@@ -0,0 +1,26 @@
+# Security Policy
+
+## Supported Versions
+
+The following table lists versions and whether they are supported. Security
+vulnerability reports will be accepted and acted upon for all supported
+versions.
+
+| Version | Supported |
+| ------- | ------------------ |
+| 0.4.x | :white_check_mark: |
+| 0.3.x | :x: |
+| < 0.3 | :x: |
+
+
+## Reporting a Vulnerability
+
+
+To report a security vulnerability, please use the [Tidelift security
+contact](https://tidelift.com/security). Tidelift will coordinate the fix and
+disclosure.
+
+If you have found a security vulnerability, in order to keep it confidential,
+please do not report an issue on GitHub.
+
+We do not award bounties for security vulnerabilities.
\ No newline at end of file
diff --git a/auto_building_tools/.circleci/config.yml b/auto_building_tools/.circleci/config.yml
new file mode 100644
index 0000000..7a98f88
--- /dev/null
+++ b/auto_building_tools/.circleci/config.yml
@@ -0,0 +1,129 @@
+version: 2.1
+
+jobs:
+ lint:
+ docker:
+ - image: cimg/python:3.9.18
+ steps:
+ - checkout
+ - run:
+ name: dependencies
+ command: |
+ source build_tools/shared.sh
+ # Include pytest compatibility with mypy
+ pip install pytest $(get_dep ruff min) $(get_dep mypy min) $(get_dep black min) cython-lint
+ - run:
+ name: linting
+ command: ./build_tools/linting.sh
+
+ doc-min-dependencies:
+ docker:
+ - image: cimg/python:3.9.18
+ environment:
+ - MKL_NUM_THREADS: 2
+ - OPENBLAS_NUM_THREADS: 2
+ - CONDA_ENV_NAME: testenv
+ - LOCK_FILE: build_tools/circle/doc_min_dependencies_linux-64_conda.lock
+ # Do not fail if the documentation build generates warnings with minimum
+ # dependencies as long as we can avoid raising warnings with more recent
+ # versions of the same dependencies.
+ - SKLEARN_WARNINGS_AS_ERRORS: '0'
+ steps:
+ - checkout
+ - run: ./build_tools/circle/checkout_merge_commit.sh
+ - restore_cache:
+ key: v1-doc-min-deps-datasets-{{ .Branch }}
+ - restore_cache:
+ keys:
+ - doc-min-deps-ccache-{{ .Branch }}
+ - doc-min-deps-ccache
+ - run: ./build_tools/circle/build_doc.sh
+ - save_cache:
+ key: doc-min-deps-ccache-{{ .Branch }}-{{ .BuildNum }}
+ paths:
+ - ~/.ccache
+ - ~/.cache/pip
+ - save_cache:
+ key: v1-doc-min-deps-datasets-{{ .Branch }}
+ paths:
+ - ~/scikit_learn_data
+ - store_artifacts:
+ path: doc/_build/html/stable
+ destination: doc
+ - store_artifacts:
+ path: ~/log.txt
+ destination: log.txt
+
+ doc:
+ docker:
+ - image: cimg/python:3.9.18
+ environment:
+ - MKL_NUM_THREADS: 2
+ - OPENBLAS_NUM_THREADS: 2
+ - CONDA_ENV_NAME: testenv
+ - LOCK_FILE: build_tools/circle/doc_linux-64_conda.lock
+ # Make sure that we fail if the documentation build generates warnings with
+ # recent versions of the dependencies.
+ - SKLEARN_WARNINGS_AS_ERRORS: '1'
+ steps:
+ - checkout
+ - run: ./build_tools/circle/checkout_merge_commit.sh
+ - restore_cache:
+ key: v1-doc-datasets-{{ .Branch }}
+ - restore_cache:
+ keys:
+ - doc-ccache-{{ .Branch }}
+ - doc-ccache
+ - run: ./build_tools/circle/build_doc.sh
+ - save_cache:
+ key: doc-ccache-{{ .Branch }}-{{ .BuildNum }}
+ paths:
+ - ~/.ccache
+ - ~/.cache/pip
+ - save_cache:
+ key: v1-doc-datasets-{{ .Branch }}
+ paths:
+ - ~/scikit_learn_data
+ - store_artifacts:
+ path: doc/_build/html/stable
+ destination: doc
+ - store_artifacts:
+ path: ~/log.txt
+ destination: log.txt
+ # Persists generated documentation so that it can be attached and deployed
+ # in the 'deploy' step.
+ - persist_to_workspace:
+ root: doc/_build/html
+ paths: .
+
+ deploy:
+ docker:
+ - image: cimg/python:3.9.18
+ steps:
+ - checkout
+ - run: ./build_tools/circle/checkout_merge_commit.sh
+ # Attach documentation generated in the 'doc' step so that it can be
+ # deployed.
+ - attach_workspace:
+ at: doc/_build/html
+ - run: ls -ltrh doc/_build/html/stable
+ - deploy:
+ command: |
+ if [[ "${CIRCLE_BRANCH}" =~ ^main$|^[0-9]+\.[0-9]+\.X$ ]]; then
+ bash build_tools/circle/push_doc.sh doc/_build/html/stable
+ fi
+
+workflows:
+ version: 2
+ build-doc-and-deploy:
+ jobs:
+ - lint
+ - doc:
+ requires:
+ - lint
+ - doc-min-dependencies:
+ requires:
+ - lint
+ - deploy:
+ requires:
+ - doc
diff --git a/auto_building_tools/.github/FUNDING.yml b/auto_building_tools/.github/FUNDING.yml
new file mode 100644
index 0000000..5662909
--- /dev/null
+++ b/auto_building_tools/.github/FUNDING.yml
@@ -0,0 +1,12 @@
+# These are supported funding model platforms
+
+github: # Replace with up to 4 GitHub Sponsors-enabled usernames e.g., [user1, user2]
+patreon: # Replace with a single Patreon username
+open_collective: # Replace with a single Open Collective username
+ko_fi: # Replace with a single Ko-fi username
+tidelift: # Replace with a single Tidelift platform-name/package-name e.g., npm/babel
+community_bridge: # Replace with a single Community Bridge project-name e.g., cloud-foundry
+liberapay: # Replace with a single Liberapay username
+issuehunt: # Replace with a single IssueHunt username
+otechie: # Replace with a single Otechie username
+custom: ['https://numfocus.org/donate-to-scikit-learn']
diff --git a/auto_building_tools/.github/ISSUE_TEMPLATE/bug_report.yml b/auto_building_tools/.github/ISSUE_TEMPLATE/bug_report.yml
new file mode 100644
index 0000000..bc8e5b5
--- /dev/null
+++ b/auto_building_tools/.github/ISSUE_TEMPLATE/bug_report.yml
@@ -0,0 +1,95 @@
+name: Bug Report
+description: Create a report to help us reproduce and correct the bug
+labels: ['Bug', 'Needs Triage']
+
+body:
+- type: markdown
+ attributes:
+ value: >
+ #### Before submitting a bug, please make sure the issue hasn't been already
+ addressed by searching through [the past issues](https://github.com/scikit-learn/scikit-learn/issues).
+- type: textarea
+ attributes:
+ label: Describe the bug
+ description: >
+ A clear and concise description of what the bug is.
+ validations:
+ required: true
+- type: textarea
+ attributes:
+ label: Steps/Code to Reproduce
+ description: |
+ Please add a [minimal code example](https://scikit-learn.org/dev/developers/minimal_reproducer.html) that can reproduce the error when running it. Be as succinct as possible, **do not depend on external data files**: instead you can generate synthetic data using `numpy.random`, [sklearn.datasets.make_regression](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.make_regression.html), [sklearn.datasets.make_classification](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.make_classification.html) or a few lines of Python code. Example:
+
+ ```python
+ from sklearn.feature_extraction.text import CountVectorizer
+ from sklearn.decomposition import LatentDirichletAllocation
+ docs = ["Help I have a bug" for i in range(1000)]
+ vectorizer = CountVectorizer(input=docs, analyzer='word')
+ lda_features = vectorizer.fit_transform(docs)
+ lda_model = LatentDirichletAllocation(
+ n_topics=10,
+ learning_method='online',
+ evaluate_every=10,
+ n_jobs=4,
+ )
+ model = lda_model.fit(lda_features)
+ ```
+
+ If the code is too long, feel free to put it in a public gist and link it in the issue: https://gist.github.com.
+
+ In short, **we are going to copy-paste your code** to run it and we expect to get the same result as you.
+
+ We acknowledge that crafting a [minimal reproducible code example](https://scikit-learn.org/dev/developers/minimal_reproducer.html) requires some effort on your side but it really helps the maintainers quickly reproduce the problem and analyze its cause without any ambiguity. Ambiguous bug reports tend to be slower to fix because they will require more effort and back and forth discussion between the maintainers and the reporter to pin-point the precise conditions necessary to reproduce the problem.
+ placeholder: |
+ ```
+ Sample code to reproduce the problem
+ ```
+ validations:
+ required: true
+- type: textarea
+ attributes:
+ label: Expected Results
+ description: >
+ Please paste or describe the expected results.
+ placeholder: >
+ Example: No error is thrown.
+ validations:
+ required: true
+- type: textarea
+ attributes:
+ label: Actual Results
+ description: |
+ Please paste or describe the results you observe instead of the expected results. If you observe an error, please paste the error message including the **full traceback** of the exception. For instance the code above raises the following exception:
+
+ ```python-traceback
+ ---------------------------------------------------------------------------
+ TypeError Traceback (most recent call last)
+  +
+\n\n```\n"
+ + log[log.find(start) + len(start) + 1 : log.find(end) - 1]
+ + "\n```\n\n
\n\n"
+ )
+ return res
+
+
+def get_message(log_file, repo, pr_number, sha, run_id, details, versions):
+ with open(log_file, "r") as f:
+ log = f.read()
+
+ sub_text = (
+ "\n\n _Generated for commit:"
+ f" [{sha[:7]}](https://github.com/{repo}/pull/{pr_number}/commits/{sha}). "
+ "Link to the linter CI: [here]"
+ f"(https://github.com/{repo}/actions/runs/{run_id})_ "
+ )
+
+ if "### Linting completed ###" not in log:
+ return (
+ "## ❌ Linting issues\n\n"
+ "There was an issue running the linter job. Please update with "
+ "`upstream/main` ([link]("
+ "https://scikit-learn.org/dev/developers/contributing.html"
+ "#how-to-contribute)) and push the changes. If you already have done "
+ "that, please send an empty commit with `git commit --allow-empty` "
+ "and push the changes to trigger the CI.\n\n" + sub_text
+ )
+
+ message = ""
+
+ # black
+ message += get_step_message(
+ log,
+ start="### Running black ###",
+ end="Problems detected by black",
+ title="`black`",
+ message=(
+ "`black` detected issues. Please run `black .` locally and push "
+ "the changes. Here you can see the detected issues. Note that "
+ "running black might also fix some of the issues which might be "
+ "detected by `ruff`. Note that the installed `black` version is "
+ f"`black={versions['black']}`."
+ ),
+ details=details,
+ )
+
+ # ruff
+ message += get_step_message(
+ log,
+ start="### Running ruff ###",
+ end="Problems detected by ruff",
+ title="`ruff`",
+ message=(
+ "`ruff` detected issues. Please run "
+ "`ruff check --fix --output-format=full .` locally, fix the remaining "
+ "issues, and push the changes. Here you can see the detected issues. Note "
+ f"that the installed `ruff` version is `ruff={versions['ruff']}`."
+ ),
+ details=details,
+ )
+
+ # mypy
+ message += get_step_message(
+ log,
+ start="### Running mypy ###",
+ end="Problems detected by mypy",
+ title="`mypy`",
+ message=(
+ "`mypy` detected issues. Please fix them locally and push the changes. "
+ "Here you can see the detected issues. Note that the installed `mypy` "
+ f"version is `mypy={versions['mypy']}`."
+ ),
+ details=details,
+ )
+
+ # cython-lint
+ message += get_step_message(
+ log,
+ start="### Running cython-lint ###",
+ end="Problems detected by cython-lint",
+ title="`cython-lint`",
+ message=(
+ "`cython-lint` detected issues. Please fix them locally and push "
+ "the changes. Here you can see the detected issues. Note that the "
+ "installed `cython-lint` version is "
+ f"`cython-lint={versions['cython-lint']}`."
+ ),
+ details=details,
+ )
+
+ # deprecation order
+ message += get_step_message(
+ log,
+ start="### Checking for bad deprecation order ###",
+ end="Problems detected by deprecation order check",
+ title="Deprecation Order",
+ message=(
+ "Deprecation order check detected issues. Please fix them locally and "
+ "push the changes. Here you can see the detected issues."
+ ),
+ details=details,
+ )
+
+ # doctest directives
+ message += get_step_message(
+ log,
+ start="### Checking for default doctest directives ###",
+ end="Problems detected by doctest directive check",
+ title="Doctest Directives",
+ message=(
+ "doctest directive check detected issues. Please fix them locally and "
+ "push the changes. Here you can see the detected issues."
+ ),
+ details=details,
+ )
+
+ # joblib imports
+ message += get_step_message(
+ log,
+ start="### Checking for joblib imports ###",
+ end="Problems detected by joblib import check",
+ title="Joblib Imports",
+ message=(
+ "`joblib` import check detected issues. Please fix them locally and "
+ "push the changes. Here you can see the detected issues."
+ ),
+ details=details,
+ )
+
+ if not message:
+ # no issues detected, so this script "fails"
+ return (
+ "## ✔️ Linting Passed\n"
+ "All linting checks passed. Your pull request is in excellent shape! ☀️"
+ + sub_text
+ )
+
+ if not details:
+ # This happens if posting the log fails, which happens if the log is too
+ # long. Typically, this happens if the PR branch hasn't been updated
+ # since we've introduced import sorting.
+ branch_not_updated = (
+ "_Merging with `upstream/main` might fix / improve the issues if you "
+ "haven't done that since 21.06.2023._\n\n"
+ )
+ else:
+ branch_not_updated = ""
+
+ message = (
+ "## ❌ Linting issues\n\n"
+ + branch_not_updated
+ + "This PR is introducing linting issues. Here's a summary of the issues. "
+ + "Note that you can avoid having linting issues by enabling `pre-commit` "
+ + "hooks. Instructions to enable them can be found [here]("
+ + "https://scikit-learn.org/dev/developers/contributing.html#how-to-contribute)"
+ + ".\n\n"
+ + "You can see the details of the linting issues under the `lint` job [here]"
+ + f"(https://github.com/{repo}/actions/runs/{run_id})\n\n"
+ + message
+ + sub_text
+ )
+
+ return message
+
+
+def get_headers(token):
+ """Get the headers for the GitHub API."""
+ return {
+ "Accept": "application/vnd.github+json",
+ "Authorization": f"Bearer {token}",
+ "X-GitHub-Api-Version": "2022-11-28",
+ }
+
+
+def find_lint_bot_comments(repo, token, pr_number):
+ """Get the comment from the linting bot."""
+ # repo is in the form of "org/repo"
+ # API doc: https://docs.github.com/en/rest/issues/comments?apiVersion=2022-11-28#list-issue-comments # noqa
+ response = requests.get(
+ f"https://api.github.com/repos/{repo}/issues/{pr_number}/comments",
+ headers=get_headers(token),
+ )
+ response.raise_for_status()
+ all_comments = response.json()
+
+ failed_comment = "❌ Linting issues"
+ success_comment = "✔️ Linting Passed"
+
+ # Find all comments that match the linting bot, and return the first one.
+ # There should always be only one such comment, or none, if the PR is
+ # just created.
+ comments = [
+ comment
+ for comment in all_comments
+ if comment["user"]["login"] == "github-actions[bot]"
+ and (failed_comment in comment["body"] or success_comment in comment["body"])
+ ]
+
+ if len(all_comments) > 25 and not comments:
+ # By default the API returns the first 30 comments. If we can't find the
+ # comment created by the bot in those, then we raise and we skip creating
+ # a comment in the first place.
+ raise RuntimeError("Comment not found in the first 30 comments.")
+
+ return comments[0] if comments else None
+
+
+def create_or_update_comment(comment, message, repo, pr_number, token):
+ """Create a new comment or update existing one."""
+ # repo is in the form of "org/repo"
+ if comment is not None:
+ print("updating existing comment")
+ # API doc: https://docs.github.com/en/rest/issues/comments?apiVersion=2022-11-28#update-an-issue-comment # noqa
+ response = requests.patch(
+ f"https://api.github.com/repos/{repo}/issues/comments/{comment['id']}",
+ headers=get_headers(token),
+ json={"body": message},
+ )
+ else:
+ print("creating new comment")
+ # API doc: https://docs.github.com/en/rest/issues/comments?apiVersion=2022-11-28#create-an-issue-comment # noqa
+ response = requests.post(
+ f"https://api.github.com/repos/{repo}/issues/{pr_number}/comments",
+ headers=get_headers(token),
+ json={"body": message},
+ )
+
+ response.raise_for_status()
+
+
+if __name__ == "__main__":
+ repo = os.environ["GITHUB_REPOSITORY"]
+ token = os.environ["GITHUB_TOKEN"]
+ pr_number = os.environ["PR_NUMBER"]
+ sha = os.environ["BRANCH_SHA"]
+ log_file = os.environ["LOG_FILE"]
+ run_id = os.environ["RUN_ID"]
+ versions_file = os.environ["VERSIONS_FILE"]
+
+ versions = get_versions(versions_file)
+
+ if not repo or not token or not pr_number or not log_file or not run_id:
+ raise ValueError(
+ "One of the following environment variables is not set: "
+ "GITHUB_REPOSITORY, GITHUB_TOKEN, PR_NUMBER, LOG_FILE, RUN_ID"
+ )
+
+ try:
+ comment = find_lint_bot_comments(repo, token, pr_number)
+ except RuntimeError:
+ print("Comment not found in the first 30 comments. Skipping!")
+ exit(0)
+
+ try:
+ message = get_message(
+ log_file,
+ repo=repo,
+ pr_number=pr_number,
+ sha=sha,
+ run_id=run_id,
+ details=True,
+ versions=versions,
+ )
+ create_or_update_comment(
+ comment=comment,
+ message=message,
+ repo=repo,
+ pr_number=pr_number,
+ token=token,
+ )
+ print(message)
+ except requests.HTTPError:
+ # The above fails if the message is too long. In that case, we
+ # try again without the details.
+ message = get_message(
+ log_file,
+ repo=repo,
+ pr_number=pr_number,
+ sha=sha,
+ run_id=run_id,
+ details=False,
+ versions=versions,
+ )
+ create_or_update_comment(
+ comment=comment,
+ message=message,
+ repo=repo,
+ pr_number=pr_number,
+ token=token,
+ )
+ print(message)
diff --git a/auto_building_tools/build_tools/github/Windows b/auto_building_tools/build_tools/github/Windows
new file mode 100644
index 0000000..a9971aa
--- /dev/null
+++ b/auto_building_tools/build_tools/github/Windows
@@ -0,0 +1,13 @@
+# Get the Python version of the base image from a build argument
+ARG PYTHON_VERSION
+FROM winamd64/python:$PYTHON_VERSION-windowsservercore
+
+ARG WHEEL_NAME
+ARG CIBW_TEST_REQUIRES
+
+# Copy and install the Windows wheel
+COPY $WHEEL_NAME $WHEEL_NAME
+RUN pip install $env:WHEEL_NAME
+
+# Install the testing dependencies
+RUN pip install $env:CIBW_TEST_REQUIRES.split(" ")
diff --git a/auto_building_tools/build_tools/github/build_minimal_windows_image.sh b/auto_building_tools/build_tools/github/build_minimal_windows_image.sh
new file mode 100644
index 0000000..2995b69
--- /dev/null
+++ b/auto_building_tools/build_tools/github/build_minimal_windows_image.sh
@@ -0,0 +1,25 @@
+#!/bin/bash
+
+set -e
+set -x
+
+PYTHON_VERSION=$1
+
+TEMP_FOLDER="$HOME/AppData/Local/Temp"
+WHEEL_PATH=$(ls -d $TEMP_FOLDER/**/*/repaired_wheel/*)
+WHEEL_NAME=$(basename $WHEEL_PATH)
+
+cp $WHEEL_PATH $WHEEL_NAME
+
+# Dot the Python version for identyfing the base Docker image
+PYTHON_VERSION=$(echo ${PYTHON_VERSION:0:1}.${PYTHON_VERSION:1:2})
+
+if [[ "$CIBW_PRERELEASE_PYTHONS" == "True" ]]; then
+ PYTHON_VERSION="$PYTHON_VERSION-rc"
+fi
+# Build a minimal Windows Docker image for testing the wheels
+docker build --build-arg PYTHON_VERSION=$PYTHON_VERSION \
+ --build-arg WHEEL_NAME=$WHEEL_NAME \
+ --build-arg CIBW_TEST_REQUIRES="$CIBW_TEST_REQUIRES" \
+ -f build_tools/github/Windows \
+ -t scikit-learn/minimal-windows .
diff --git a/auto_building_tools/build_tools/github/build_source.sh b/auto_building_tools/build_tools/github/build_source.sh
new file mode 100644
index 0000000..ec53284
--- /dev/null
+++ b/auto_building_tools/build_tools/github/build_source.sh
@@ -0,0 +1,20 @@
+#!/bin/bash
+
+set -e
+set -x
+
+# Move up two levels to create the virtual
+# environment outside of the source folder
+cd ../../
+
+python -m venv build_env
+source build_env/bin/activate
+
+python -m pip install numpy scipy cython
+python -m pip install twine build
+
+cd scikit-learn/scikit-learn
+python -m build --sdist
+
+# Check whether the source distribution will render correctly
+twine check dist/*.tar.gz
diff --git a/auto_building_tools/build_tools/github/check_build_trigger.sh b/auto_building_tools/build_tools/github/check_build_trigger.sh
new file mode 100644
index 0000000..e3a02c4
--- /dev/null
+++ b/auto_building_tools/build_tools/github/check_build_trigger.sh
@@ -0,0 +1,14 @@
+#!/bin/bash
+
+set -e
+set -x
+
+COMMIT_MSG=$(git log --no-merges -1 --oneline)
+
+# The commit marker "[cd build]" or "[cd build gh]" will trigger the build when required
+if [[ "$GITHUB_EVENT_NAME" == schedule ||
+ "$GITHUB_EVENT_NAME" == workflow_dispatch ||
+ "$COMMIT_MSG" =~ \[cd\ build\] ||
+ "$COMMIT_MSG" =~ \[cd\ build\ gh\] ]]; then
+ echo "build=true" >> $GITHUB_OUTPUT
+fi
diff --git a/auto_building_tools/build_tools/github/check_wheels.py b/auto_building_tools/build_tools/github/check_wheels.py
new file mode 100644
index 0000000..5579d86
--- /dev/null
+++ b/auto_building_tools/build_tools/github/check_wheels.py
@@ -0,0 +1,37 @@
+"""Checks that dist/* contains the number of wheels built from the
+.github/workflows/wheels.yml config."""
+
+import sys
+from pathlib import Path
+
+import yaml
+
+gh_wheel_path = Path.cwd() / ".github" / "workflows" / "wheels.yml"
+with gh_wheel_path.open("r") as f:
+ wheel_config = yaml.safe_load(f)
+
+build_matrix = wheel_config["jobs"]["build_wheels"]["strategy"]["matrix"]["include"]
+n_wheels = len(build_matrix)
+
+# plus one more for the sdist
+n_wheels += 1
+
+# arm64 builds from cirrus
+cirrus_path = Path.cwd() / "build_tools" / "cirrus" / "arm_wheel.yml"
+with cirrus_path.open("r") as f:
+ cirrus_config = yaml.safe_load(f)
+
+n_wheels += len(cirrus_config["linux_arm64_wheel_task"]["matrix"])
+
+dist_files = list(Path("dist").glob("**/*"))
+n_dist_files = len(dist_files)
+
+if n_dist_files != n_wheels:
+ print(
+ f"Expected {n_wheels} wheels in dist/* but "
+ f"got {n_dist_files} artifacts instead."
+ )
+ sys.exit(1)
+
+print(f"dist/* has the expected {n_wheels} wheels:")
+print("\n".join(file.name for file in dist_files))
diff --git a/auto_building_tools/build_tools/github/create_gpu_environment.sh b/auto_building_tools/build_tools/github/create_gpu_environment.sh
new file mode 100644
index 0000000..96a62d7
--- /dev/null
+++ b/auto_building_tools/build_tools/github/create_gpu_environment.sh
@@ -0,0 +1,20 @@
+#!/bin/bash
+
+set -e
+set -x
+
+curl -L -O "https://github.com/conda-forge/miniforge/releases/latest/download/Miniforge3-$(uname)-$(uname -m).sh"
+bash Miniforge3-$(uname)-$(uname -m).sh -b -p "${HOME}/conda"
+source "${HOME}/conda/etc/profile.d/conda.sh"
+
+
+# defines the get_dep and show_installed_libraries functions
+source build_tools/shared.sh
+conda activate base
+
+CONDA_ENV_NAME=sklearn
+LOCK_FILE=build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_conda.lock
+create_conda_environment_from_lock_file $CONDA_ENV_NAME $LOCK_FILE
+
+conda activate $CONDA_ENV_NAME
+conda list
diff --git a/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_conda.lock b/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_conda.lock
new file mode 100644
index 0000000..08c24ad
--- /dev/null
+++ b/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_conda.lock
@@ -0,0 +1,261 @@
+# Generated by conda-lock.
+# platform: linux-64
+# input_hash: 7044e24fc9243a244c265e4b8c44e1304a8f55cd0cfa2d036ead6f92921d624e
+@EXPLICIT
+https://conda.anaconda.org/conda-forge/linux-64/_libgcc_mutex-0.1-conda_forge.tar.bz2#d7c89558ba9fa0495403155b64376d81
+https://conda.anaconda.org/conda-forge/linux-64/ca-certificates-2024.7.4-hbcca054_0.conda#23ab7665c5f63cfb9f1f6195256daac6
+https://conda.anaconda.org/conda-forge/noarch/cuda-version-12.4-h3060b56_3.conda#c9a3fe8b957176e1a8452c6f3431b0d8
+https://conda.anaconda.org/conda-forge/noarch/font-ttf-dejavu-sans-mono-2.37-hab24e00_0.tar.bz2#0c96522c6bdaed4b1566d11387caaf45
+https://conda.anaconda.org/conda-forge/noarch/font-ttf-inconsolata-3.000-h77eed37_0.tar.bz2#34893075a5c9e55cdafac56607368fc6
+https://conda.anaconda.org/conda-forge/noarch/font-ttf-source-code-pro-2.038-h77eed37_0.tar.bz2#4d59c254e01d9cde7957100457e2d5fb
+https://conda.anaconda.org/conda-forge/noarch/font-ttf-ubuntu-0.83-h77eed37_2.conda#cbbe59391138ea5ad3658c76912e147f
+https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.40-hf3520f5_7.conda#b80f2f396ca2c28b8c14c437a4ed1e74
+https://conda.anaconda.org/conda-forge/linux-64/mkl-include-2022.1.0-h84fe81f_915.tar.bz2#2dcd1acca05c11410d4494d7fc7dfa2a
+https://conda.anaconda.org/conda-forge/linux-64/python_abi-3.12-5_cp312.conda#0424ae29b104430108f5218a66db7260
+https://conda.anaconda.org/pytorch/noarch/pytorch-mutex-1.0-cuda.tar.bz2#a948316e36fb5b11223b3fcfa93f8358
+https://conda.anaconda.org/conda-forge/noarch/tzdata-2024a-h0c530f3_0.conda#161081fc7cec0bfda0d86d7cb595f8d8
+https://conda.anaconda.org/conda-forge/noarch/cuda-cccl_linux-64-12.4.127-ha770c72_2.conda#357fbcd43f1296b02d6738a2295abc24
+https://conda.anaconda.org/conda-forge/noarch/cuda-cudart-static_linux-64-12.4.127-h85509e4_2.conda#0b0522d8685968f25370d5c36bb9fba3
+https://conda.anaconda.org/conda-forge/noarch/cuda-cudart_linux-64-12.4.127-h85509e4_2.conda#329163110a96514802e9e64d971edf43
+https://conda.anaconda.org/conda-forge/noarch/fonts-conda-forge-1-0.tar.bz2#f766549260d6815b0c52253f1fb1bb29
+https://conda.anaconda.org/conda-forge/linux-64/libglvnd-1.7.0-ha4b6fd6_0.conda#e46b5ae31282252e0525713e34ffbe2b
+https://conda.anaconda.org/conda-forge/noarch/cuda-cudart-dev_linux-64-12.4.127-h85509e4_2.conda#12039deb2a3f103f5756831702bf29fc
+https://conda.anaconda.org/conda-forge/noarch/fonts-conda-ecosystem-1-0.tar.bz2#fee5683a3f04bd15cbd8318b096a27ab
+https://conda.anaconda.org/conda-forge/linux-64/libegl-1.7.0-ha4b6fd6_0.conda#35e52d19547cb3265a09c49de146a5ae
+https://conda.anaconda.org/conda-forge/linux-64/_openmp_mutex-4.5-2_kmp_llvm.tar.bz2#562b26ba2e19059551a811e72ab7f793
+https://conda.anaconda.org/conda-forge/linux-64/libgcc-ng-14.1.0-h77fa898_0.conda#ca0fad6a41ddaef54a153b78eccb5037
+https://conda.anaconda.org/conda-forge/linux-64/alsa-lib-1.2.12-h4ab18f5_0.conda#7ed427f0871fd41cb1d9c17727c17589
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-common-0.9.27-h4bc722e_0.conda#817119e8a21a45d325f65d0d54710052
+https://conda.anaconda.org/conda-forge/linux-64/bzip2-1.0.8-h4bc722e_7.conda#62ee74e96c5ebb0af99386de58cf9553
+https://conda.anaconda.org/conda-forge/linux-64/c-ares-1.33.1-heb4867d_0.conda#0d3c60291342c0c025db231353376dfb
+https://conda.anaconda.org/conda-forge/linux-64/keyutils-1.6.1-h166bdaf_0.tar.bz2#30186d27e2c9fa62b45fb1476b7200e3
+https://conda.anaconda.org/conda-forge/linux-64/libbrotlicommon-1.1.0-hd590300_1.conda#aec6c91c7371c26392a06708a73c70e5
+https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.21-h4bc722e_0.conda#36ce76665bf67f5aac36be7a0d21b7f3
+https://conda.anaconda.org/conda-forge/linux-64/libev-4.33-hd590300_2.conda#172bf1cd1ff8629f2b1179945ed45055
+https://conda.anaconda.org/conda-forge/linux-64/libexpat-2.6.2-h59595ed_0.conda#e7ba12deb7020dd080c6c70e7b6f6a3d
+https://conda.anaconda.org/conda-forge/linux-64/libffi-3.4.2-h7f98852_5.tar.bz2#d645c6d2ac96843a2bfaccd2d62b3ac3
+https://conda.anaconda.org/conda-forge/linux-64/libgfortran5-14.1.0-hc5f4f2c_0.conda#6456c2620c990cd8dde2428a27ba0bc5
+https://conda.anaconda.org/conda-forge/linux-64/libiconv-1.17-hd590300_2.conda#d66573916ffcf376178462f1b61c941e
+https://conda.anaconda.org/conda-forge/linux-64/libjpeg-turbo-3.0.0-hd590300_1.conda#ea25936bb4080d843790b586850f82b8
+https://conda.anaconda.org/conda-forge/linux-64/libnsl-2.0.1-hd590300_0.conda#30fd6e37fe21f86f4bd26d6ee73eeec7
+https://conda.anaconda.org/conda-forge/linux-64/libpciaccess-0.18-hd590300_0.conda#48f4330bfcd959c3cfb704d424903c82
+https://conda.anaconda.org/conda-forge/linux-64/libstdcxx-ng-14.1.0-hc0a3c3a_0.conda#1cb187a157136398ddbaae90713e2498
+https://conda.anaconda.org/conda-forge/linux-64/libutf8proc-2.8.0-h166bdaf_0.tar.bz2#ede4266dc02e875fe1ea77b25dd43747
+https://conda.anaconda.org/conda-forge/linux-64/libuuid-2.38.1-h0b41bf4_0.conda#40b61aab5c7ba9ff276c41cfffe6b80b
+https://conda.anaconda.org/conda-forge/linux-64/libwebp-base-1.4.0-hd590300_0.conda#b26e8aa824079e1be0294e7152ca4559
+https://conda.anaconda.org/conda-forge/linux-64/libxcrypt-4.4.36-hd590300_1.conda#5aa797f8787fe7a17d1b0821485b5adc
+https://conda.anaconda.org/conda-forge/linux-64/libzlib-1.3.1-h4ab18f5_1.conda#57d7dc60e9325e3de37ff8dffd18e814
+https://conda.anaconda.org/conda-forge/linux-64/ncurses-6.5-h59595ed_0.conda#fcea371545eda051b6deafb24889fc69
+https://conda.anaconda.org/conda-forge/linux-64/ocl-icd-2.3.2-hd590300_1.conda#c66f837ac65e4d1cdeb80e2a1d5fcc3d
+https://conda.anaconda.org/conda-forge/linux-64/openssl-3.3.1-hb9d3cd8_3.conda#6c566a46baae794daf34775d41eb180a
+https://conda.anaconda.org/conda-forge/linux-64/pthread-stubs-0.4-h36c2ea0_1001.tar.bz2#22dad4df6e8630e8dff2428f6f6a7036
+https://conda.anaconda.org/conda-forge/linux-64/xorg-inputproto-2.3.2-h7f98852_1002.tar.bz2#bcd1b3396ec6960cbc1d2855a9e60b2b
+https://conda.anaconda.org/conda-forge/linux-64/xorg-kbproto-1.0.7-h7f98852_1002.tar.bz2#4b230e8381279d76131116660f5a241a
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libice-1.1.1-hd590300_0.conda#b462a33c0be1421532f28bfe8f4a7514
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxau-1.0.11-hd590300_0.conda#2c80dc38fface310c9bd81b17037fee5
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxdmcp-1.1.3-h7f98852_0.tar.bz2#be93aabceefa2fac576e971aef407908
+https://conda.anaconda.org/conda-forge/linux-64/xorg-recordproto-1.14.2-h7f98852_1002.tar.bz2#2f835e6c386e73c6faaddfe9eda67e98
+https://conda.anaconda.org/conda-forge/linux-64/xorg-renderproto-0.11.1-h7f98852_1002.tar.bz2#06feff3d2634e3097ce2fe681474b534
+https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h0b41bf4_1003.conda#bce9f945da8ad2ae9b1d7165a64d0f87
+https://conda.anaconda.org/conda-forge/linux-64/xorg-xproto-7.0.31-h7f98852_1007.tar.bz2#b4a4381d54784606820704f7b5f05a15
+https://conda.anaconda.org/conda-forge/linux-64/xz-5.2.6-h166bdaf_0.tar.bz2#2161070d867d1b1204ea749c8eec4ef0
+https://conda.anaconda.org/conda-forge/linux-64/yaml-0.2.5-h7f98852_2.tar.bz2#4cb3ad778ec2d5a7acbdf254eb1c42ae
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-cal-0.7.3-h8dac057_2.conda#577509458a061ddc9b089602ac6e1e98
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-compression-0.2.19-haa50ccc_0.conda#00c38c49d0befb632f686cf67ee8c9f5
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-sdkutils-0.1.19-h038f3f9_2.conda#6861cab6cddb5d713cb3db95c838d30f
+https://conda.anaconda.org/conda-forge/linux-64/aws-checksums-0.1.18-h038f3f9_10.conda#4bf9c8fcf2bb6793c55e5c5758b9b011
+https://conda.anaconda.org/conda-forge/linux-64/cuda-cudart-12.4.127-he02047a_2.conda#a748faa52331983fc3adcc3b116fe0e4
+https://conda.anaconda.org/conda-forge/linux-64/cuda-cupti-12.4.127-he02047a_2.conda#46422ef1b1161fb180027e50c598ecd0
+https://conda.anaconda.org/conda-forge/linux-64/cuda-nvrtc-12.4.127-he02047a_2.conda#80baf6262f4a1a0dde42d85aaa393402
+https://conda.anaconda.org/conda-forge/linux-64/cuda-nvtx-12.4.127-he02047a_2.conda#656a004b6e44f50ce71c65cab0d429b4
+https://conda.anaconda.org/conda-forge/linux-64/cuda-opencl-12.4.127-he02047a_1.conda#1e98deda07c14d26c80d124cf0eb011a
+https://conda.anaconda.org/conda-forge/linux-64/double-conversion-3.3.0-h59595ed_0.conda#c2f83a5ddadadcdb08fe05863295ee97
+https://conda.anaconda.org/conda-forge/linux-64/expat-2.6.2-h59595ed_0.conda#53fb86322bdb89496d7579fe3f02fd61
+https://conda.anaconda.org/conda-forge/linux-64/gflags-2.2.2-he1b5a44_1004.tar.bz2#cddaf2c63ea4a5901cf09524c490ecdc
+https://conda.anaconda.org/conda-forge/linux-64/gmp-6.3.0-hac33072_2.conda#c94a5994ef49749880a8139cf9afcbe1
+https://conda.anaconda.org/conda-forge/linux-64/graphite2-1.3.13-h59595ed_1003.conda#f87c7b7c2cb45f323ffbce941c78ab7c
+https://conda.anaconda.org/conda-forge/linux-64/icu-75.1-he02047a_0.conda#8b189310083baabfb622af68fd9d3ae3
+https://conda.anaconda.org/conda-forge/linux-64/lerc-4.0.0-h27087fc_0.tar.bz2#76bbff344f0134279f225174e9064c8f
+https://conda.anaconda.org/conda-forge/linux-64/libabseil-20240116.2-cxx17_he02047a_1.conda#c48fc56ec03229f294176923c3265c05
+https://conda.anaconda.org/conda-forge/linux-64/libbrotlidec-1.1.0-hd590300_1.conda#f07002e225d7a60a694d42a7bf5ff53f
+https://conda.anaconda.org/conda-forge/linux-64/libbrotlienc-1.1.0-hd590300_1.conda#5fc11c6020d421960607d821310fcd4d
+https://conda.anaconda.org/conda-forge/linux-64/libcrc32c-1.1.2-h9c3ff4c_0.tar.bz2#c965a5aa0d5c1c37ffc62dff36e28400
+https://conda.anaconda.org/conda-forge/linux-64/libcufft-11.2.0.44-hd3aeb46_0.conda#dc9bd2796334ff4779aacccc53221b31
+https://conda.anaconda.org/conda-forge/linux-64/libcufile-1.9.1.3-he02047a_2.conda#a051267bcb1912467c81d802a7d3465e
+https://conda.anaconda.org/conda-forge/linux-64/libcurand-10.3.5.147-he02047a_2.conda#9c4886d513fd477df88d411cd274c202
+https://conda.anaconda.org/conda-forge/linux-64/libdrm-2.4.122-h4ab18f5_0.conda#bbfc4dbe5e97b385ef088f354d65e563
+https://conda.anaconda.org/conda-forge/linux-64/libedit-3.1.20191231-he28a2e2_2.tar.bz2#4d331e44109e3f0e19b4cb8f9b82f3e1
+https://conda.anaconda.org/conda-forge/linux-64/libevent-2.1.12-hf998b51_1.conda#a1cfcc585f0c42bf8d5546bb1dfb668d
+https://conda.anaconda.org/conda-forge/linux-64/libgfortran-ng-14.1.0-h69a702a_0.conda#f4ca84fbd6d06b0a052fb2d5b96dde41
+https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.58.0-h47da74e_1.conda#700ac6ea6d53d5510591c4344d5c989a
+https://conda.anaconda.org/conda-forge/linux-64/libnpp-12.2.5.2-hd3aeb46_0.conda#3bd78f0d576dfa2af0588371cfe662dc
+https://conda.anaconda.org/conda-forge/linux-64/libnvfatbin-12.4.127-he02047a_2.conda#d746b76642b4ac6e40f1219405672beb
+https://conda.anaconda.org/conda-forge/linux-64/libnvjitlink-12.4.99-hd3aeb46_0.conda#2121026ab1e941622333f4a047d69fa5
+https://conda.anaconda.org/conda-forge/linux-64/libnvjpeg-12.3.1.89-h59595ed_0.conda#f2ef6f1215587a366c56b462dcb6e1fe
+https://conda.anaconda.org/conda-forge/linux-64/libpng-1.6.43-h2797004_0.conda#009981dd9cfcaa4dbfa25ffaed86bcae
+https://conda.anaconda.org/conda-forge/linux-64/libsqlite-3.46.0-hde9e2c9_0.conda#18aa975d2094c34aef978060ae7da7d8
+https://conda.anaconda.org/conda-forge/linux-64/libssh2-1.11.0-h0841786_0.conda#1f5a58e686b13bcfde88b93f547d23fe
+https://conda.anaconda.org/conda-forge/linux-64/libxcb-1.16-hb9d3cd8_1.conda#3601598f0db0470af28985e3e7ad0158
+https://conda.anaconda.org/conda-forge/linux-64/llvm-openmp-15.0.7-h0cdce71_0.conda#589c9a3575a050b583241c3d688ad9aa
+https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.4-hcb278e6_0.conda#318b08df404f9c9be5712aaa5a6f0bb0
+https://conda.anaconda.org/conda-forge/linux-64/mysql-common-9.0.1-h70512c7_0.conda#c567b6fa201bc424e84f1e70f7a36095
+https://conda.anaconda.org/conda-forge/linux-64/ninja-1.12.1-h297d8ca_0.conda#3aa1c7e292afeff25a0091ddd7c69b72
+https://conda.anaconda.org/conda-forge/linux-64/pcre2-10.44-hba22ea6_2.conda#df359c09c41cd186fffb93a2d87aa6f5
+https://conda.anaconda.org/conda-forge/linux-64/pixman-0.43.2-h59595ed_0.conda#71004cbf7924e19c02746ccde9fd7123
+https://conda.anaconda.org/conda-forge/linux-64/qhull-2020.2-h434a139_5.conda#353823361b1d27eb3960efb076dfcaf6
+https://conda.anaconda.org/conda-forge/linux-64/readline-8.2-h8228510_1.conda#47d31b792659ce70f470b5c82fdfb7a4
+https://conda.anaconda.org/conda-forge/linux-64/s2n-1.5.1-h3400bea_0.conda#bf136eb7f8e15fcf8915c1a04b0aec6f
+https://conda.anaconda.org/conda-forge/linux-64/snappy-1.2.1-ha2e4443_0.conda#6b7dcc7349efd123d493d2dbe85a045f
+https://conda.anaconda.org/conda-forge/linux-64/tk-8.6.13-noxft_h4845f30_101.conda#d453b98d9c83e71da0741bb0ff4d76bc
+https://conda.anaconda.org/conda-forge/linux-64/wayland-1.23.1-h3e06ad9_0.conda#0a732427643ae5e0486a727927791da1
+https://conda.anaconda.org/conda-forge/linux-64/xorg-fixesproto-5.0-h7f98852_1002.tar.bz2#65ad6e1eb4aed2b0611855aff05e04f6
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libsm-1.2.4-h7391055_0.conda#93ee23f12bc2e684548181256edd2cf6
+https://conda.anaconda.org/conda-forge/linux-64/zlib-1.3.1-h4ab18f5_1.conda#9653f1bf3766164d0e65fa723cabbc54
+https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.6-ha6fb4c9_0.conda#4d056880988120e29d75bfff282e0f45
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-io-0.14.18-hf5b9b93_6.conda#8fd43c2719355d795f5c7cef11f08ec0
+https://conda.anaconda.org/conda-forge/linux-64/brotli-bin-1.1.0-hd590300_1.conda#39f910d205726805a958da408ca194ba
+https://conda.anaconda.org/conda-forge/linux-64/freetype-2.12.1-h267a509_2.conda#9ae35c3d96db2c94ce0cef86efdfa2cb
+https://conda.anaconda.org/conda-forge/linux-64/glog-0.7.1-hbabe93e_0.conda#ff862eebdfeb2fd048ae9dc92510baca
+https://conda.anaconda.org/conda-forge/linux-64/krb5-1.21.3-h659f571_0.conda#3f43953b7d3fb3aaa1d0d0723d91e368
+https://conda.anaconda.org/conda-forge/linux-64/libcublas-12.4.2.65-hd3aeb46_0.conda#a83c33c2abd3bd467e13329a9655cb03
+https://conda.anaconda.org/conda-forge/linux-64/libcusparse-12.3.0.142-hd3aeb46_0.conda#d72fd369532e32f800ab6447e9cd00ac
+https://conda.anaconda.org/conda-forge/linux-64/libglib-2.80.3-h315aac3_2.conda#b0143a3e98136a680b728fdf9b42a258
+https://conda.anaconda.org/conda-forge/linux-64/libhiredis-1.0.2-h2cc385e_0.tar.bz2#b34907d3a81a3cd8095ee83d174c074a
+https://conda.anaconda.org/conda-forge/linux-64/libprotobuf-4.25.3-h08a7969_0.conda#6945825cebd2aeb16af4c69d97c32c13
+https://conda.anaconda.org/conda-forge/linux-64/libre2-11-2023.09.01-h5a48ba9_2.conda#41c69fba59d495e8cf5ffda48a607e35
+https://conda.anaconda.org/conda-forge/linux-64/libthrift-0.20.0-hb90f79a_0.conda#9ce07c1750e779c9d4cc968047f78b0d
+https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.6.0-h46a8edc_4.conda#a7e3a62981350e232e0e7345b5aea580
+https://conda.anaconda.org/conda-forge/linux-64/libxml2-2.12.7-he7c6b58_4.conda#08a9265c637230c37cb1be4a6cad4536
+https://conda.anaconda.org/conda-forge/linux-64/mpfr-4.2.1-h38ae2d0_2.conda#168e18a2bba4f8520e6c5e38982f5847
+https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-9.0.1-ha479ceb_0.conda#6fd406aef37faad86bd7f37a94fb6f8a
+https://conda.anaconda.org/conda-forge/linux-64/python-3.12.5-h2ad013b_0_cpython.conda#9c56c4df45f6571b13111d8df2448692
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-0.4.1-hb711507_2.conda#8637c3e5821654d0edf97e2b0404b443
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-keysyms-0.4.1-hb711507_0.conda#ad748ccca349aec3e91743e08b5e2b50
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-renderutil-0.3.10-hb711507_0.conda#0e0cbe0564d03a99afd5fd7b362feecd
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-wm-0.4.2-hb711507_0.conda#608e0ef8256b81d04456e8d211eee3e8
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.8.9-hb711507_1.conda#4a6d410296d7e39f00bacdee7df046e9
+https://conda.anaconda.org/conda-forge/noarch/array-api-compat-1.8-pyhd8ed1ab_0.conda#1178a75b8f6f260ac4b4436979754278
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-event-stream-0.4.3-h570d160_0.conda#1c121949295cac86798be8f369768d7c
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-http-0.8.7-h1c59cda_5.conda#0fc88e5bb5f095bdf4129282411c50c9
+https://conda.anaconda.org/conda-forge/linux-64/brotli-1.1.0-hd590300_1.conda#f27a24d46e3ea7b70a1f98e50c62508f
+https://conda.anaconda.org/conda-forge/linux-64/ccache-4.10.1-h065aff2_0.conda#d6b48c138e0c8170a6fe9c136e063540
+https://conda.anaconda.org/conda-forge/noarch/certifi-2024.7.4-pyhd8ed1ab_0.conda#24e7fd6ca65997938fff9e5ab6f653e4
+https://conda.anaconda.org/conda-forge/noarch/colorama-0.4.6-pyhd8ed1ab_0.tar.bz2#3faab06a954c2a04039983f2c4a50d99
+https://conda.anaconda.org/conda-forge/noarch/cycler-0.12.1-pyhd8ed1ab_0.conda#5cd86562580f274031ede6aa6aa24441
+https://conda.anaconda.org/conda-forge/linux-64/cython-3.0.11-py312hca68cad_0.conda#f824c60def49466ad5b9aed4eaa23c28
+https://conda.anaconda.org/conda-forge/linux-64/dbus-1.13.6-h5008d03_3.tar.bz2#ecfff944ba3960ecb334b9a2663d708d
+https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.2.2-pyhd8ed1ab_0.conda#d02ae936e42063ca46af6cdad2dbd1e0
+https://conda.anaconda.org/conda-forge/noarch/execnet-2.1.1-pyhd8ed1ab_0.conda#15dda3cdbf330abfe9f555d22f66db46
+https://conda.anaconda.org/conda-forge/linux-64/fastrlock-0.8.2-py312h30efb56_2.conda#7065ec5a4909f925e305b77e505b0aec
+https://conda.anaconda.org/conda-forge/noarch/filelock-3.15.4-pyhd8ed1ab_0.conda#0e7e4388e9d5283e22b35a9443bdbcc9
+https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.2-h14ed4e7_0.conda#0f69b688f52ff6da70bccb7ff7001d1d
+https://conda.anaconda.org/conda-forge/noarch/iniconfig-2.0.0-pyhd8ed1ab_0.conda#f800d2da156d08e289b14e87e43c1ae5
+https://conda.anaconda.org/conda-forge/linux-64/kiwisolver-1.4.5-py312h8572e83_1.conda#c1e71f2bc05d8e8e033aefac2c490d05
+https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.16-hb7c19ff_0.conda#51bb7010fc86f70eee639b4bb7a894f5
+https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h4637d8d_4.conda#d4529f4dff3057982a7617c7ac58fde3
+https://conda.anaconda.org/conda-forge/linux-64/libcurl-8.9.1-hdb1bdb2_0.conda#7da1d242ca3591e174a3c7d82230d3c0
+https://conda.anaconda.org/conda-forge/linux-64/libcusolver-11.6.0.99-hd3aeb46_0.conda#ccea7d03d84947a00296f6a39dc774b8
+https://conda.anaconda.org/conda-forge/linux-64/libglx-1.7.0-ha4b6fd6_0.conda#b470cc353c5b852e0d830e8d5d23e952
+https://conda.anaconda.org/conda-forge/linux-64/libhwloc-2.11.1-default_hecaa2ac_1000.conda#f54aeebefb5c5ff84eca4fb05ca8aa3a
+https://conda.anaconda.org/conda-forge/linux-64/libllvm18-18.1.8-h8b73ec9_2.conda#2e25bb2f53e4a48873a936f8ef53e592
+https://conda.anaconda.org/conda-forge/linux-64/libpq-16.4-h482b261_0.conda#0f74c5581623f860e7baca042d9d7139
+https://conda.anaconda.org/conda-forge/linux-64/libxslt-1.1.39-h76b75d6_0.conda#e71f31f8cfb0a91439f2086fc8aa0461
+https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.5-py312h98912ed_0.conda#6ff0b9582da2d4a74a1f9ae1f9ce2af6
+https://conda.anaconda.org/conda-forge/linux-64/mpc-1.3.1-hfe3b2da_0.conda#289c71e83dc0daa7d4c81f04180778ca
+https://conda.anaconda.org/conda-forge/noarch/mpmath-1.3.0-pyhd8ed1ab_0.conda#dbf6e2d89137da32fa6670f3bffc024e
+https://conda.anaconda.org/conda-forge/noarch/munkres-1.1.4-pyh9f0ad1d_0.tar.bz2#2ba8498c1018c1e9c61eb99b973dfe19
+https://conda.anaconda.org/conda-forge/noarch/networkx-3.3-pyhd8ed1ab_1.conda#d335fd5704b46f4efb89a6774e81aef0
+https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.2-h488ebb8_0.conda#7f2e286780f072ed750df46dc2631138
+https://conda.anaconda.org/conda-forge/linux-64/orc-2.0.2-h669347b_0.conda#1e6c10f7d749a490612404efeb179eb8
+https://conda.anaconda.org/conda-forge/noarch/packaging-24.1-pyhd8ed1ab_0.conda#cbe1bb1f21567018ce595d9c2be0f0db
+https://conda.anaconda.org/conda-forge/noarch/pluggy-1.5.0-pyhd8ed1ab_0.conda#d3483c8fc2dc2cc3f5cf43e26d60cabf
+https://conda.anaconda.org/conda-forge/noarch/pyparsing-3.1.4-pyhd8ed1ab_0.conda#4d91352a50949d049cf9714c8563d433
+https://conda.anaconda.org/conda-forge/noarch/python-tzdata-2024.1-pyhd8ed1ab_0.conda#98206ea9954216ee7540f0c773f2104d
+https://conda.anaconda.org/conda-forge/noarch/pytz-2024.1-pyhd8ed1ab_0.conda#3eeeeb9e4827ace8c0c1419c85d590ad
+https://conda.anaconda.org/conda-forge/linux-64/pyyaml-6.0.2-py312h41a817b_0.conda#1779c9cbd9006415ab7bb9e12747e9d1
+https://conda.anaconda.org/conda-forge/linux-64/re2-2023.09.01-h7f4b329_2.conda#8f70e36268dea8eb666ef14c29bd3cda
+https://conda.anaconda.org/conda-forge/noarch/setuptools-72.2.0-pyhd8ed1ab_0.conda#1462aa8b243aad09ef5d0841c745eb89
+https://conda.anaconda.org/conda-forge/noarch/six-1.16.0-pyh6c4a22f_0.tar.bz2#e5f25f8dbc060e9a8d912e432202afc2
+https://conda.anaconda.org/conda-forge/noarch/threadpoolctl-3.5.0-pyhc1e730c_0.conda#df68d78237980a159bd7149f33c0e8fd
+https://conda.anaconda.org/conda-forge/noarch/toml-0.10.2-pyhd8ed1ab_0.tar.bz2#f832c45a477c78bebd107098db465095
+https://conda.anaconda.org/conda-forge/noarch/tomli-2.0.1-pyhd8ed1ab_0.tar.bz2#5844808ffab9ebdb694585b50ba02a96
+https://conda.anaconda.org/conda-forge/linux-64/tornado-6.4.1-py312h9a8786e_0.conda#fd9c83fde763b494f07acee1404c280e
+https://conda.anaconda.org/conda-forge/noarch/typing_extensions-4.12.2-pyha770c72_0.conda#ebe6952715e1d5eb567eeebf25250fa7
+https://conda.anaconda.org/conda-forge/noarch/wheel-0.44.0-pyhd8ed1ab_0.conda#d44e3b085abcaef02983c6305b84b584
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-image-0.4.0-hb711507_2.conda#a0901183f08b6c7107aab109733a3c91
+https://conda.anaconda.org/conda-forge/linux-64/xkeyboard-config-2.42-h4ab18f5_0.conda#b193af204da1bfb8c13882d131a14bd2
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h0b41bf4_2.conda#82b6df12252e6f32402b96dacc656fec
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxfixes-5.0.3-h7f98852_1004.tar.bz2#e9a21aa4d5e3e5f1aed71e8cefd46b6a
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxrender-0.9.11-hd590300_0.conda#ed67c36f215b310412b2af935bf3e530
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-auth-0.7.25-h15d0e8c_6.conda#e0d292ba383ac09598c664186c0144cd
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-mqtt-0.10.4-hc14a930_17.conda#f0e3f95a9f545d5975e8573f80cdb5fa
+https://conda.anaconda.org/conda-forge/linux-64/azure-core-cpp-1.13.0-h935415a_0.conda#debd1677c2fea41eb2233a260f48a298
+https://conda.anaconda.org/conda-forge/linux-64/cairo-1.18.0-hebfffa5_3.conda#fceaedf1cdbcb02df9699a0d9b005292
+https://conda.anaconda.org/conda-forge/linux-64/coverage-7.6.1-py312h41a817b_0.conda#4006636c39312dc42f8504475be3800f
+https://conda.anaconda.org/nvidia/linux-64/cuda-libraries-12.4.0-0.tar.bz2#e1f3474ec98d3a4e17d791389c07e769
+https://conda.anaconda.org/conda-forge/linux-64/fonttools-4.53.1-py312h41a817b_0.conda#da921c56bcf69a8b97216ecec0cc4015
+https://conda.anaconda.org/conda-forge/linux-64/gmpy2-2.1.5-py312h1d5cde6_1.conda#27abd7664bc87595bd98b6306b8393d1
+https://conda.anaconda.org/conda-forge/noarch/jinja2-3.1.4-pyhd8ed1ab_0.conda#7b86ecb7d3557821c649b3c31e3eb9f2
+https://conda.anaconda.org/conda-forge/noarch/joblib-1.4.2-pyhd8ed1ab_0.conda#25df261d4523d9f9783bcdb7208d872f
+https://conda.anaconda.org/conda-forge/linux-64/libclang-cpp18.1-18.1.8-default_hf981a13_2.conda#b0f8c590aa86d9bee5987082f7f15bdf
+https://conda.anaconda.org/conda-forge/linux-64/libclang13-18.1.8-default_h9def88c_2.conda#ba2d12adbea9de311297f2b577f4bb86
+https://conda.anaconda.org/conda-forge/linux-64/libgl-1.7.0-ha4b6fd6_0.conda#3deca8c25851196c28d1c84dd4ae9149
+https://conda.anaconda.org/conda-forge/linux-64/libgrpc-1.62.2-h15f2491_0.conda#8dabe607748cb3d7002ad73cd06f1325
+https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.7.0-h2c5496b_1.conda#e2eaefa4de2b7237af7c907b8bbc760a
+https://conda.anaconda.org/conda-forge/noarch/meson-1.5.1-pyhd8ed1ab_1.conda#979087ee59bea1355f991a3b738af64e
+https://conda.anaconda.org/conda-forge/linux-64/pillow-10.4.0-py312h287a98d_0.conda#59ea71eed98aee0bebbbdd3b118167c7
+https://conda.anaconda.org/conda-forge/noarch/pip-24.2-pyhd8ed1ab_0.conda#6721aef6bfe5937abe70181545dd2c51
+https://conda.anaconda.org/conda-forge/noarch/pyproject-metadata-0.8.0-pyhd8ed1ab_0.conda#573fe09d7bd0cd4bcc210d8369b5ca47
+https://conda.anaconda.org/conda-forge/noarch/pytest-8.3.2-pyhd8ed1ab_0.conda#e010a224b90f1f623a917c35addbb924
+https://conda.anaconda.org/conda-forge/noarch/python-dateutil-2.9.0-pyhd8ed1ab_0.conda#2cf4264fffb9e6eff6031c5b6884d61c
+https://conda.anaconda.org/conda-forge/linux-64/tbb-2021.12.0-h434a139_3.conda#c667c11d1e488a38220ede8a34441bff
+https://conda.anaconda.org/conda-forge/linux-64/xcb-util-cursor-0.1.4-h4ab18f5_2.conda#79e46d4a6ccecb7ee1912042958a8758
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxi-1.7.10-h4bc722e_1.conda#749baebe7e2ff3360630e069175e528b
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxxf86vm-1.1.5-h4bc722e_1.conda#0c90ad87101001080484b91bd9d2cdef
+https://conda.anaconda.org/conda-forge/linux-64/aws-c-s3-0.6.4-h558cea2_8.conda#af03e7b03e929396fb80ffac1a676c89
+https://conda.anaconda.org/conda-forge/linux-64/azure-identity-cpp-1.8.0-hd126650_2.conda#36df3cf05459de5d0a41c77c4329634b
+https://conda.anaconda.org/conda-forge/linux-64/azure-storage-common-cpp-12.7.0-h10ac4d7_1.conda#ab6d507ad16dbe2157920451d662e4a1
+https://conda.anaconda.org/conda-forge/noarch/cuda-runtime-12.4.0-ha804496_0.conda#b760ac3b8e6faaf4f59cb2c47334b4f3
+https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-9.0.0-hda332d3_1.conda#76b32dcf243444aea9c6b804bcfa40b8
+https://conda.anaconda.org/conda-forge/linux-64/libgoogle-cloud-2.28.0-h26d7fe4_0.conda#2c51703b4d775f8943c08a361788131b
+https://conda.anaconda.org/conda-forge/noarch/meson-python-0.16.0-pyh0c530f3_0.conda#e16f0dbf502da873be9f9adb0dc52547
+https://conda.anaconda.org/conda-forge/linux-64/mkl-2022.1.0-h84fe81f_915.tar.bz2#b9c8f925797a93dbff45e1626b025a6b
+https://conda.anaconda.org/conda-forge/noarch/pytest-cov-5.0.0-pyhd8ed1ab_0.conda#c54c0107057d67ddf077751339ec2c63
+https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.6.1-pyhd8ed1ab_0.conda#b39568655c127a9c4a44d178ac99b6d0
+https://conda.anaconda.org/conda-forge/noarch/sympy-1.13.2-pypyh2585a3b_103.conda#7327125b427c98b81564f164c4a75d4c
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxtst-1.2.5-h4bc722e_0.conda#185159d666308204eca00295599b0a5c
+https://conda.anaconda.org/conda-forge/linux-64/aws-crt-cpp-0.27.6-h1966bd9_0.conda#30b59fa809914489974fe275a0fb7c7e
+https://conda.anaconda.org/conda-forge/linux-64/azure-storage-blobs-cpp-12.12.0-hd2e3451_0.conda#61f1c193452f0daa582f39634627ea33
+https://conda.anaconda.org/conda-forge/linux-64/libblas-3.9.0-16_linux64_mkl.tar.bz2#85f61af03fd291dae33150ffe89dc09a
+https://conda.anaconda.org/conda-forge/linux-64/libgoogle-cloud-storage-2.28.0-ha262f82_0.conda#9e7960f0b9ab3895ef73d92477c47dae
+https://conda.anaconda.org/conda-forge/linux-64/mkl-devel-2022.1.0-ha770c72_916.tar.bz2#69ba49e445f87aea2cba343a71a35ca2
+https://conda.anaconda.org/pytorch/linux-64/pytorch-cuda-12.4-hc786d27_6.tar.bz2#294df2aee019b0e314713842d46e6b65
+https://conda.anaconda.org/conda-forge/linux-64/qt6-main-6.7.2-hb12f9c5_5.conda#8c662388c2418f293266f5e7f50df7d7
+https://conda.anaconda.org/conda-forge/linux-64/aws-sdk-cpp-1.11.379-hf9693f6_5.conda#18a4bf7e8a65006b26ca53700fcf2362
+https://conda.anaconda.org/conda-forge/linux-64/azure-storage-files-datalake-cpp-12.11.0-h325d260_1.conda#11d926d1f4a75a1b03d1c053ca20424b
+https://conda.anaconda.org/conda-forge/linux-64/libcblas-3.9.0-16_linux64_mkl.tar.bz2#361bf757b95488de76c4f123805742d3
+https://conda.anaconda.org/conda-forge/linux-64/liblapack-3.9.0-16_linux64_mkl.tar.bz2#a2f166748917d6d6e4707841ca1f519e
+https://conda.anaconda.org/conda-forge/linux-64/pyside6-6.7.2-py312hb5137db_2.conda#99889d0c042cc4dfb9a758619d487282
+https://conda.anaconda.org/conda-forge/linux-64/libarrow-17.0.0-h9d17f36_9_cpu.conda#bfae79329f50d5bd960e1ac289625096
+https://conda.anaconda.org/conda-forge/linux-64/liblapacke-3.9.0-16_linux64_mkl.tar.bz2#44ccc4d4dca6a8d57fa17442bc64b5a1
+https://conda.anaconda.org/conda-forge/linux-64/numpy-2.1.0-py312h1103770_0.conda#9709027e8a51a3476db65a3c0cf806c2
+https://conda.anaconda.org/conda-forge/noarch/array-api-strict-2.0.1-pyhd8ed1ab_0.conda#2c00d29e0e276f2d32dfe20e698b8eeb
+https://conda.anaconda.org/conda-forge/linux-64/blas-devel-3.9.0-16_linux64_mkl.tar.bz2#3f92c1c9e1c0e183462c5071aa02cae1
+https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.2.1-py312h8572e83_0.conda#12c6a831ef734f0b2dd4caff514cbb7f
+https://conda.anaconda.org/conda-forge/linux-64/cupy-core-13.3.0-py312h28031eb_0.conda#e3c07012b06106c20120e2a8f8d9e79c
+https://conda.anaconda.org/conda-forge/linux-64/libarrow-acero-17.0.0-h5888daf_9_cpu.conda#cace9fe91c532c67ff828937a633fb1c
+https://conda.anaconda.org/conda-forge/linux-64/libparquet-17.0.0-h39682fd_9_cpu.conda#0efe4b18e72f519298f57ff75a9adf07
+https://conda.anaconda.org/conda-forge/linux-64/pandas-2.2.2-py312h1d6d2e6_1.conda#ae00b61f3000d2284d1f2584d4dfafa8
+https://conda.anaconda.org/conda-forge/linux-64/polars-1.5.0-py312h7285250_0.conda#4756b2dda06b6c7bedb376677ffbca06
+https://conda.anaconda.org/conda-forge/linux-64/pyarrow-core-17.0.0-py312h9cafe31_1_cpu.conda#235827b9c93850cafdd2d5ab359893f9
+https://conda.anaconda.org/conda-forge/linux-64/scipy-1.14.1-py312h7d485d2_0.conda#7418a22e73008356d9aba99d93dfeeee
+https://conda.anaconda.org/conda-forge/linux-64/blas-2.116-mkl.tar.bz2#c196a26abf6b4f132c88828ab7c2231c
+https://conda.anaconda.org/conda-forge/linux-64/cupy-13.3.0-py312h7d319b9_0.conda#eff11ea4f19a2b52080d317d9a38fe0a
+https://conda.anaconda.org/conda-forge/linux-64/libarrow-dataset-17.0.0-h5888daf_9_cpu.conda#4df21168065a9e21372a442783dfd547
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.9.2-py312h854627b_0.conda#a57b0ae7c0aac603839a4e83a3e997d6
+https://conda.anaconda.org/conda-forge/linux-64/pyamg-5.2.1-py312h389efb2_0.conda#37038b979f8be9666d90a852879368fb
+https://conda.anaconda.org/conda-forge/linux-64/libarrow-substrait-17.0.0-hf54134d_9_cpu.conda#239401053cfbf93d24795b12dec89c56
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.9.2-py312h7900ff3_0.conda#44c07eccf73f549b8ea5c9aacfe3ad0a
+https://conda.anaconda.org/conda-forge/linux-64/pyarrow-17.0.0-py312h9cebb41_1.conda#7e8ddbd44fb99ba376b09c4e9e61e509
+https://conda.anaconda.org/pytorch/linux-64/pytorch-2.4.0-py3.12_cuda12.4_cudnn9.1.0_0.tar.bz2#9731ae9086ed66acc02e8e4aba5d9990
+https://conda.anaconda.org/pytorch/linux-64/torchtriton-3.0.0-py312.tar.bz2#e53c6345daef28009cd51187a5c5af73
diff --git a/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_environment.yml b/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_environment.yml
new file mode 100644
index 0000000..e2ffb14
--- /dev/null
+++ b/auto_building_tools/build_tools/github/pylatest_conda_forge_cuda_array-api_linux-64_environment.yml
@@ -0,0 +1,34 @@
+# DO NOT EDIT: this file is generated from the specification found in the
+# following script to centralize the configuration for CI builds:
+# build_tools/update_environments_and_lock_files.py
+channels:
+ - conda-forge
+ - pytorch
+ - nvidia
+dependencies:
+ - python
+ - numpy
+ - blas
+ - scipy
+ - cython
+ - joblib
+ - threadpoolctl
+ - matplotlib
+ - pandas
+ - pyamg
+ - pytest
+ - pytest-xdist
+ - pillow
+ - pip
+ - ninja
+ - meson-python
+ - pytest-cov
+ - coverage
+ - ccache
+ - pytorch::pytorch
+ - pytorch-cuda
+ - polars
+ - pyarrow
+ - cupy
+ - array-api-compat
+ - array-api-strict
diff --git a/auto_building_tools/build_tools/github/repair_windows_wheels.sh b/auto_building_tools/build_tools/github/repair_windows_wheels.sh
new file mode 100644
index 0000000..8f51a34
--- /dev/null
+++ b/auto_building_tools/build_tools/github/repair_windows_wheels.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+
+set -e
+set -x
+
+WHEEL=$1
+DEST_DIR=$2
+
+# By default, the Windows wheels are not repaired.
+# In this case, we need to vendor VCRUNTIME140.dll
+pip install wheel
+wheel unpack "$WHEEL"
+WHEEL_DIRNAME=$(ls -d scikit_learn-*)
+python build_tools/github/vendor.py "$WHEEL_DIRNAME"
+wheel pack "$WHEEL_DIRNAME" -d "$DEST_DIR"
+rm -rf "$WHEEL_DIRNAME"
diff --git a/auto_building_tools/build_tools/github/test_source.sh b/auto_building_tools/build_tools/github/test_source.sh
new file mode 100644
index 0000000..c93d22a
--- /dev/null
+++ b/auto_building_tools/build_tools/github/test_source.sh
@@ -0,0 +1,18 @@
+#!/bin/bash
+
+set -e
+set -x
+
+cd ../../
+
+python -m venv test_env
+source test_env/bin/activate
+
+python -m pip install scikit-learn/scikit-learn/dist/*.tar.gz
+python -m pip install pytest pandas
+
+# Run the tests on the installed source distribution
+mkdir tmp_for_test
+cd tmp_for_test
+
+pytest --pyargs sklearn
diff --git a/auto_building_tools/build_tools/github/test_windows_wheels.sh b/auto_building_tools/build_tools/github/test_windows_wheels.sh
new file mode 100644
index 0000000..07954a7
--- /dev/null
+++ b/auto_building_tools/build_tools/github/test_windows_wheels.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+
+set -e
+set -x
+
+PYTHON_VERSION=$1
+
+docker container run \
+ --rm scikit-learn/minimal-windows \
+ powershell -Command "python -c 'import sklearn; sklearn.show_versions()'"
+
+docker container run \
+ -e SKLEARN_SKIP_NETWORK_TESTS=1 \
+ --rm scikit-learn/minimal-windows \
+ powershell -Command "pytest --pyargs sklearn"
diff --git a/auto_building_tools/build_tools/github/upload_anaconda.sh b/auto_building_tools/build_tools/github/upload_anaconda.sh
new file mode 100644
index 0000000..51401dd
--- /dev/null
+++ b/auto_building_tools/build_tools/github/upload_anaconda.sh
@@ -0,0 +1,24 @@
+#!/bin/bash
+
+set -e
+set -x
+
+if [[ "$GITHUB_EVENT_NAME" == "schedule" \
+ || "$GITHUB_EVENT_NAME" == "workflow_dispatch" \
+ || "$CIRRUS_CRON" == "nightly" ]]; then
+ ANACONDA_ORG="scientific-python-nightly-wheels"
+ ANACONDA_TOKEN="$SCIKIT_LEARN_NIGHTLY_UPLOAD_TOKEN"
+else
+ ANACONDA_ORG="scikit-learn-wheels-staging"
+ ANACONDA_TOKEN="$SCIKIT_LEARN_STAGING_UPLOAD_TOKEN"
+fi
+
+# Install Python 3.8 because of a bug with Python 3.9
+export PATH=$CONDA/bin:$PATH
+conda create -n upload -y python=3.8
+source activate upload
+conda install -y anaconda-client
+
+# Force a replacement if the remote file already exists
+anaconda -t $ANACONDA_TOKEN upload --force -u $ANACONDA_ORG $ARTIFACTS_PATH/*
+echo "Index: https://pypi.anaconda.org/$ANACONDA_ORG/simple"
diff --git a/auto_building_tools/build_tools/github/vendor.py b/auto_building_tools/build_tools/github/vendor.py
new file mode 100644
index 0000000..28b44be
--- /dev/null
+++ b/auto_building_tools/build_tools/github/vendor.py
@@ -0,0 +1,96 @@
+"""Embed vcomp140.dll and msvcp140.dll."""
+
+import os
+import os.path as op
+import shutil
+import sys
+import textwrap
+
+TARGET_FOLDER = op.join("sklearn", ".libs")
+DISTRIBUTOR_INIT = op.join("sklearn", "_distributor_init.py")
+VCOMP140_SRC_PATH = "C:\\Windows\\System32\\vcomp140.dll"
+MSVCP140_SRC_PATH = "C:\\Windows\\System32\\msvcp140.dll"
+
+
+def make_distributor_init_64_bits(
+ distributor_init,
+ vcomp140_dll_filename,
+ msvcp140_dll_filename,
+):
+ """Create a _distributor_init.py file for 64-bit architectures.
+
+ This file is imported first when importing the sklearn package
+ so as to pre-load the vendored vcomp140.dll and msvcp140.dll.
+ """
+ with open(distributor_init, "wt") as f:
+ f.write(
+ textwrap.dedent(
+ """
+ '''Helper to preload vcomp140.dll and msvcp140.dll to prevent
+ "not found" errors.
+
+ Once vcomp140.dll and msvcp140.dll are
+ preloaded, the namespace is made available to any subsequent
+ vcomp140.dll and msvcp140.dll. This is
+ created as part of the scripts that build the wheel.
+ '''
+
+
+ import os
+ import os.path as op
+ from ctypes import WinDLL
+
+
+ if os.name == "nt":
+ libs_path = op.join(op.dirname(__file__), ".libs")
+ vcomp140_dll_filename = op.join(libs_path, "{0}")
+ msvcp140_dll_filename = op.join(libs_path, "{1}")
+ WinDLL(op.abspath(vcomp140_dll_filename))
+ WinDLL(op.abspath(msvcp140_dll_filename))
+ """.format(
+ vcomp140_dll_filename,
+ msvcp140_dll_filename,
+ )
+ )
+ )
+
+
+def main(wheel_dirname):
+ """Embed vcomp140.dll and msvcp140.dll."""
+ if not op.exists(VCOMP140_SRC_PATH):
+ raise ValueError(f"Could not find {VCOMP140_SRC_PATH}.")
+
+ if not op.exists(MSVCP140_SRC_PATH):
+ raise ValueError(f"Could not find {MSVCP140_SRC_PATH}.")
+
+ if not op.isdir(wheel_dirname):
+ raise RuntimeError(f"Could not find {wheel_dirname} file.")
+
+ vcomp140_dll_filename = op.basename(VCOMP140_SRC_PATH)
+ msvcp140_dll_filename = op.basename(MSVCP140_SRC_PATH)
+
+ target_folder = op.join(wheel_dirname, TARGET_FOLDER)
+ distributor_init = op.join(wheel_dirname, DISTRIBUTOR_INIT)
+
+ # Create the "sklearn/.libs" subfolder
+ if not op.exists(target_folder):
+ os.mkdir(target_folder)
+
+ print(f"Copying {VCOMP140_SRC_PATH} to {target_folder}.")
+ shutil.copy2(VCOMP140_SRC_PATH, target_folder)
+
+ print(f"Copying {MSVCP140_SRC_PATH} to {target_folder}.")
+ shutil.copy2(MSVCP140_SRC_PATH, target_folder)
+
+ # Generate the _distributor_init file in the source tree
+ print("Generating the '_distributor_init.py' file.")
+ make_distributor_init_64_bits(
+ distributor_init,
+ vcomp140_dll_filename,
+ msvcp140_dll_filename,
+ )
+
+
+if __name__ == "__main__":
+ _, wheel_file = sys.argv
+ main(wheel_file)
diff --git a/auto_building_tools/build_tools/linting.sh b/auto_building_tools/build_tools/linting.sh
new file mode 100644
index 0000000..aefabfa
--- /dev/null
+++ b/auto_building_tools/build_tools/linting.sh
@@ -0,0 +1,125 @@
+#!/bin/bash
+
+# Note that any change in this file, adding or removing steps or changing the
+# printed messages, should be also reflected in the `get_comment.py` file.
+
+# This script shouldn't exit if a command / pipeline fails
+set +e
+# pipefail is necessary to propagate exit codes
+set -o pipefail
+
+global_status=0
+
+echo -e "### Running black ###\n"
+black --check --diff .
+status=$?
+
+if [[ $status -eq 0 ]]
+then
+ echo -e "No problem detected by black\n"
+else
+ echo -e "Problems detected by black, please run black and commit the result\n"
+ global_status=1
+fi
+
+echo -e "### Running ruff ###\n"
+ruff check --output-format=full .
+status=$?
+if [[ $status -eq 0 ]]
+then
+ echo -e "No problem detected by ruff\n"
+else
+ echo -e "Problems detected by ruff, please fix them\n"
+ global_status=1
+fi
+
+echo -e "### Running mypy ###\n"
+mypy sklearn/
+status=$?
+if [[ $status -eq 0 ]]
+then
+ echo -e "No problem detected by mypy\n"
+else
+ echo -e "Problems detected by mypy, please fix them\n"
+ global_status=1
+fi
+
+echo -e "### Running cython-lint ###\n"
+cython-lint sklearn/
+status=$?
+if [[ $status -eq 0 ]]
+then
+ echo -e "No problem detected by cython-lint\n"
+else
+ echo -e "Problems detected by cython-lint, please fix them\n"
+ global_status=1
+fi
+
+# For docstrings and warnings of deprecated attributes to be rendered
+# properly, the `deprecated` decorator must come before the `property` decorator
+# (else they are treated as functions)
+
+echo -e "### Checking for bad deprecation order ###\n"
+bad_deprecation_property_order=`git grep -A 10 "@property" -- "*.py" | awk '/@property/,/def /' | grep -B1 "@deprecated"`
+
+if [ ! -z "$bad_deprecation_property_order" ]
+then
+ echo "deprecated decorator should come before property decorator"
+ echo "found the following occurrences:"
+ echo $bad_deprecation_property_order
+ echo -e "\nProblems detected by deprecation order check\n"
+ global_status=1
+else
+ echo -e "No problems detected related to deprecation order\n"
+fi
+
+# Check for default doctest directives ELLIPSIS and NORMALIZE_WHITESPACE
+
+echo -e "### Checking for default doctest directives ###\n"
+doctest_directive="$(git grep -nw -E "# doctest\: \+(ELLIPSIS|NORMALIZE_WHITESPACE)")"
+
+if [ ! -z "$doctest_directive" ]
+then
+ echo "ELLIPSIS and NORMALIZE_WHITESPACE doctest directives are enabled by default, but were found in:"
+ echo "$doctest_directive"
+ echo -e "\nProblems detected by doctest directive check\n"
+ global_status=1
+else
+ echo -e "No problems detected related to doctest directives\n"
+fi
+
+# Check for joblib.delayed and joblib.Parallel imports
+# TODO(1.7): remove ":!sklearn/utils/_joblib.py"
+echo -e "### Checking for joblib imports ###\n"
+joblib_status=0
+joblib_delayed_import="$(git grep -l -A 10 -E "joblib import.+delayed" -- "*.py" ":!sklearn/utils/_joblib.py" ":!sklearn/utils/parallel.py")"
+if [ ! -z "$joblib_delayed_import" ]; then
+ echo "Use from sklearn.utils.parallel import delayed instead of joblib delayed. The following files contains imports to joblib.delayed:"
+ echo "$joblib_delayed_import"
+ joblib_status=1
+fi
+joblib_Parallel_import="$(git grep -l -A 10 -E "joblib import.+Parallel" -- "*.py" ":!sklearn/utils/_joblib.py" ":!sklearn/utils/parallel.py")"
+if [ ! -z "$joblib_Parallel_import" ]; then
+ echo "Use from sklearn.utils.parallel import Parallel instead of joblib Parallel. The following files contains imports to joblib.Parallel:"
+ echo "$joblib_Parallel_import"
+ joblib_status=1
+fi
+
+if [[ $joblib_status -eq 0 ]]
+then
+ echo -e "No problems detected related to joblib imports\n"
+else
+ echo -e "\nProblems detected by joblib import check\n"
+ global_status=1
+fi
+
+echo -e "### Linting completed ###\n"
+
+if [[ $global_status -eq 1 ]]
+then
+ echo -e "Linting failed\n"
+ exit 1
+else
+ echo -e "Linting passed\n"
+ exit 0
+fi
diff --git a/auto_building_tools/build_tools/on_pr_comment_update_environments_and_lock_files.py b/auto_building_tools/build_tools/on_pr_comment_update_environments_and_lock_files.py
new file mode 100644
index 0000000..f57a17a
--- /dev/null
+++ b/auto_building_tools/build_tools/on_pr_comment_update_environments_and_lock_files.py
@@ -0,0 +1,73 @@
+import argparse
+import os
+import shlex
+import subprocess
+
+
+def execute_command(command):
+ command_list = shlex.split(command)
+ subprocess.run(command_list, check=True, text=True)
+
+
+def main():
+ comment = os.environ["COMMENT"].splitlines()[0].strip()
+
+ # Extract the command-line arguments from the comment
+ prefix = "@scikit-learn-bot update lock-files"
+ assert comment.startswith(prefix)
+ all_args_list = shlex.split(comment[len(prefix) :])
+
+ # Parse the options for the lock-file script
+ parser = argparse.ArgumentParser()
+ parser.add_argument("--select-build", default="")
+ parser.add_argument("--skip-build", default=None)
+ parser.add_argument("--select-tag", default=None)
+ args, extra_args_list = parser.parse_known_args(all_args_list)
+
+ # Rebuild the command-line arguments for the lock-file script
+ args_string = ""
+ if args.select_build != "":
+ args_string += f" --select-build {args.select_build}"
+ if args.skip_build is not None:
+ args_string += f" --skip-build {args.skip_build}"
+ if args.select_tag is not None:
+ args_string += f" --select-tag {args.select_tag}"
+
+ # Parse extra arguments
+ extra_parser = argparse.ArgumentParser()
+ extra_parser.add_argument("--commit-marker", default=None)
+ extra_args, _ = extra_parser.parse_known_args(extra_args_list)
+
+ marker = ""
+ # Additional markers based on the tag
+ if args.select_tag == "main-ci":
+ marker += "[doc build] "
+ elif args.select_tag == "scipy-dev":
+ marker += "[scipy-dev] "
+ elif args.select_tag == "arm":
+ marker += "[cirrus arm] "
+ elif len(all_args_list) == 0:
+ # No arguments which will update all lock files so add all markers
+ marker += "[doc build] [scipy-dev] [cirrus arm] "
+ # The additional `--commit-marker` argument
+ if extra_args.commit_marker is not None:
+ marker += extra_args.commit_marker + " "
+
+ execute_command(
+ f"python build_tools/update_environments_and_lock_files.py{args_string}"
+ )
+ execute_command('git config --global user.name "scikit-learn-bot"')
+ execute_command('git config --global user.email "noreply@github.com"')
+ execute_command("git add -A")
+ # Avoiding commiting the scripts that are downloaded from main
+ execute_command("git reset build_tools/shared.sh")
+ execute_command("git reset build_tools/update_environments_and_lock_files.py")
+ execute_command(
+ "git reset build_tools/on_pr_comment_update_environments_and_lock_files.py"
+ )
+ execute_command(f'git commit -m "{marker}Update lock files"')
+ execute_command("git push")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/auto_building_tools/build_tools/shared.sh b/auto_building_tools/build_tools/shared.sh
new file mode 100644
index 0000000..cb52422
--- /dev/null
+++ b/auto_building_tools/build_tools/shared.sh
@@ -0,0 +1,51 @@
+get_dep() {
+ package="$1"
+ version="$2"
+ if [[ "$version" == "none" ]]; then
+ # do not install with none
+ echo
+ elif [[ "${version%%[^0-9.]*}" ]]; then
+ # version number is explicitly passed
+ echo "$package==$version"
+ elif [[ "$version" == "latest" ]]; then
+ # use latest
+ echo "$package"
+ elif [[ "$version" == "min" ]]; then
+ echo "$package==$(python sklearn/_min_dependencies.py $package)"
+ fi
+}
+
+show_installed_libraries(){
+ # use conda list when inside a conda environment. conda list shows more
+ # info than pip list, e.g. whether OpenBLAS or MKL is installed as well as
+ # the version of OpenBLAS or MKL
+ if [[ -n "$CONDA_PREFIX" ]]; then
+ conda list
+ else
+ python -m pip list
+ fi
+}
+
+activate_environment() {
+ if [[ "$DISTRIB" =~ ^conda.* ]]; then
+ source activate $VIRTUALENV
+ elif [[ "$DISTRIB" == "ubuntu" || "$DISTRIB" == "debian-32" || "$DISTRIB" == "pip-free-threaded" ]]; then
+ source $VIRTUALENV/bin/activate
+ fi
+}
+
+create_conda_environment_from_lock_file() {
+ ENV_NAME=$1
+ LOCK_FILE=$2
+ # Because we are using lock-files with the "explicit" format, conda can
+ # install them directly, provided the lock-file does not contain pip solved
+ # packages. For more details, see
+ # https://conda.github.io/conda-lock/output/#explicit-lockfile
+ lock_file_has_pip_packages=$(grep -q files.pythonhosted.org $LOCK_FILE && echo "true" || echo "false")
+ if [[ "$lock_file_has_pip_packages" == "false" ]]; then
+ conda create --name $ENV_NAME --file $LOCK_FILE
+ else
+ conda install "$(get_dep conda-lock min)" -y
+ conda-lock install --name $ENV_NAME $LOCK_FILE
+ fi
+}
diff --git a/auto_building_tools/build_tools/update_environments_and_lock_files.py b/auto_building_tools/build_tools/update_environments_and_lock_files.py
new file mode 100644
index 0000000..2d6b6db
--- /dev/null
+++ b/auto_building_tools/build_tools/update_environments_and_lock_files.py
@@ -0,0 +1,767 @@
+"""Script to update CI environment files and associated lock files.
+
+To run it you need to be in the root folder of the scikit-learn repo:
+python build_tools/update_environments_and_lock_files.py
+
+Two scenarios where this script can be useful:
+- make sure that the latest versions of all the dependencies are used in the CI.
+ There is a scheduled workflow that does this, see
+ .github/workflows/update-lock-files.yml. This is still useful to run this
+ script when when the automated PR fails and for example some packages need to
+ be pinned. You can add the pins to this script, run it, and open a PR with
+ the changes.
+- bump minimum dependencies in sklearn/_min_dependencies.py. Running this
+ script will update both the CI environment files and associated lock files.
+ You can then open a PR with the changes.
+- pin some packages to an older version by adding them to the
+ default_package_constraints variable. This is useful when regressions are
+ introduced in our dependencies, this has happened for example with pytest 7
+ and coverage 6.3.
+
+Environments are conda environment.yml or pip requirements.txt. Lock files are
+conda-lock lock files or pip-compile requirements.txt.
+
+pip requirements.txt are used when we install some dependencies (e.g. numpy and
+scipy) with apt-get and the rest of the dependencies (e.g. pytest and joblib)
+with pip.
+
+To run this script you need:
+- conda-lock. The version should match the one used in the CI in
+ sklearn/_min_dependencies.py
+- pip-tools
+
+To only update the environment and lock files for specific builds, you can use
+the command line argument `--select-build` which will take a regex. For example,
+to only update the documentation builds you can use:
+`python build_tools/update_environments_and_lock_files.py --select-build doc`
+"""
+
+import json
+import logging
+import re
+import subprocess
+import sys
+from importlib.metadata import version
+from pathlib import Path
+
+import click
+from jinja2 import Environment
+from packaging.version import Version
+
+logger = logging.getLogger(__name__)
+logger.setLevel(logging.INFO)
+handler = logging.StreamHandler()
+logger.addHandler(handler)
+
+TRACE = logging.DEBUG - 5
+
+
+common_dependencies_without_coverage = [
+ "python",
+ "numpy",
+ "blas",
+ "scipy",
+ "cython",
+ "joblib",
+ "threadpoolctl",
+ "matplotlib",
+ "pandas",
+ "pyamg",
+ "pytest",
+ "pytest-xdist",
+ "pillow",
+ "pip",
+ "ninja",
+ "meson-python",
+]
+
+common_dependencies = common_dependencies_without_coverage + [
+ "pytest-cov",
+ "coverage",
+]
+
+docstring_test_dependencies = ["sphinx", "numpydoc"]
+
+default_package_constraints = {}
+
+
+def remove_from(alist, to_remove):
+ return [each for each in alist if each not in to_remove]
+
+
+build_metadata_list = [
+ {
+ "name": "pylatest_conda_forge_cuda_array-api_linux-64",
+ "type": "conda",
+ "tag": "cuda",
+ "folder": "build_tools/github",
+ "platform": "linux-64",
+ "channels": ["conda-forge", "pytorch", "nvidia"],
+ "conda_dependencies": common_dependencies
+ + [
+ "ccache",
+ # Make sure pytorch comes from the pytorch channel and not conda-forge
+ "pytorch::pytorch",
+ "pytorch-cuda",
+ "polars",
+ "pyarrow",
+ "cupy",
+ "array-api-compat",
+ "array-api-strict",
+ ],
+ },
+ {
+ "name": "pylatest_conda_forge_mkl_linux-64",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "linux-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": common_dependencies
+ + [
+ "ccache",
+ "pytorch",
+ "pytorch-cpu",
+ "polars",
+ "pyarrow",
+ "array-api-compat",
+ "array-api-strict",
+ ],
+ "package_constraints": {
+ "blas": "[build=mkl]",
+ },
+ },
+ {
+ "name": "pylatest_conda_forge_mkl_osx-64",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "osx-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": common_dependencies
+ + [
+ "ccache",
+ "compilers",
+ "llvm-openmp",
+ ],
+ "package_constraints": {
+ "blas": "[build=mkl]",
+ },
+ },
+ {
+ "name": "pylatest_conda_mkl_no_openmp",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "osx-64",
+ "channels": ["defaults"],
+ "conda_dependencies": remove_from(
+ common_dependencies, ["cython", "threadpoolctl", "meson-python"]
+ )
+ + ["ccache"],
+ "package_constraints": {
+ "blas": "[build=mkl]",
+ # scipy 1.12.x crashes on this platform (https://github.com/scipy/scipy/pull/20086)
+ # TODO: release scipy constraint when 1.13 is available in the "default"
+ # channel.
+ "scipy": "<1.12",
+ # TODO temporary to avoid a timeout in the no-OpenMP build, see
+ # https://github.com/scikit-learn/scikit-learn/pull/29486#issuecomment-2242359516
+ "meson": "<1.5",
+ },
+ # TODO: put cython, threadpoolctl and meson-python back to conda
+ # dependencies when required version is available on the main channel
+ "pip_dependencies": ["cython", "threadpoolctl", "meson-python", "meson"],
+ },
+ {
+ "name": "pymin_conda_forge_openblas_min_dependencies",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "linux-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": common_dependencies + ["ccache", "polars"],
+ "package_constraints": {
+ "python": "3.9",
+ "blas": "[build=openblas]",
+ "numpy": "min",
+ "scipy": "min",
+ "matplotlib": "min",
+ "cython": "min",
+ "joblib": "min",
+ "threadpoolctl": "min",
+ "meson-python": "min",
+ "pandas": "min",
+ "polars": "min",
+ },
+ },
+ {
+ "name": "pymin_conda_forge_openblas_ubuntu_2204",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "linux-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": (
+ common_dependencies_without_coverage
+ + docstring_test_dependencies
+ + ["ccache"]
+ ),
+ "package_constraints": {
+ "python": "3.9",
+ "blas": "[build=openblas]",
+ },
+ },
+ {
+ "name": "pylatest_pip_openblas_pandas",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "linux-64",
+ "channels": ["defaults"],
+ "conda_dependencies": ["python", "ccache"],
+ "pip_dependencies": (
+ remove_from(common_dependencies, ["python", "blas", "pip"])
+ + docstring_test_dependencies
+ # Test with some optional dependencies
+ + ["lightgbm", "scikit-image"]
+ # Test array API on CPU without PyTorch
+ + ["array-api-compat", "array-api-strict"]
+ ),
+ "package_constraints": {
+ # XXX: we would like to use the latest Python version, but for now using
+ # Python 3.12 makes the CI much slower so we use Python 3.11. See
+ # https://github.com/scikit-learn/scikit-learn/pull/29444#issuecomment-2219550662.
+ "python": "3.11",
+ },
+ },
+ {
+ "name": "pylatest_pip_scipy_dev",
+ "type": "conda",
+ "tag": "scipy-dev",
+ "folder": "build_tools/azure",
+ "platform": "linux-64",
+ "channels": ["defaults"],
+ "conda_dependencies": ["python", "ccache"],
+ "pip_dependencies": (
+ remove_from(
+ common_dependencies,
+ [
+ "python",
+ "blas",
+ "matplotlib",
+ "pyamg",
+ # all the dependencies below have a development version
+ # installed in the CI, so they can be removed from the
+ # environment.yml
+ "numpy",
+ "scipy",
+ "pandas",
+ "cython",
+ "joblib",
+ "pillow",
+ ],
+ )
+ + ["pooch"]
+ + docstring_test_dependencies
+ # python-dateutil is a dependency of pandas and pandas is removed from
+ # the environment.yml. Adding python-dateutil so it is pinned
+ + ["python-dateutil"]
+ ),
+ },
+ {
+ "name": "pymin_conda_forge_mkl",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "platform": "win-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": remove_from(common_dependencies, ["pandas", "pyamg"])
+ + [
+ "wheel",
+ "pip",
+ ],
+ "package_constraints": {
+ "python": "3.9",
+ "blas": "[build=mkl]",
+ },
+ },
+ {
+ "name": "doc_min_dependencies",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/circle",
+ "platform": "linux-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": common_dependencies_without_coverage
+ + [
+ "scikit-image",
+ "seaborn",
+ "memory_profiler",
+ "compilers",
+ "sphinx",
+ "sphinx-gallery",
+ "sphinx-copybutton",
+ "numpydoc",
+ "sphinx-prompt",
+ "plotly",
+ "polars",
+ "pooch",
+ "sphinx-remove-toctrees",
+ "sphinx-design",
+ "pydata-sphinx-theme",
+ ],
+ "pip_dependencies": [
+ "sphinxext-opengraph",
+ "sphinxcontrib-sass",
+ ],
+ "package_constraints": {
+ "python": "3.9",
+ "numpy": "min",
+ "scipy": "min",
+ "matplotlib": "min",
+ "cython": "min",
+ "scikit-image": "min",
+ "sphinx": "min",
+ "pandas": "min",
+ "sphinx-gallery": "min",
+ "sphinx-copybutton": "min",
+ "numpydoc": "min",
+ "sphinx-prompt": "min",
+ "sphinxext-opengraph": "min",
+ "plotly": "min",
+ "polars": "min",
+ "pooch": "min",
+ "sphinx-design": "min",
+ "sphinxcontrib-sass": "min",
+ "sphinx-remove-toctrees": "min",
+ "pydata-sphinx-theme": "min",
+ },
+ },
+ {
+ "name": "doc",
+ "type": "conda",
+ "tag": "main-ci",
+ "folder": "build_tools/circle",
+ "platform": "linux-64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": common_dependencies_without_coverage
+ + [
+ "scikit-image",
+ "seaborn",
+ "memory_profiler",
+ "compilers",
+ "sphinx",
+ "sphinx-gallery",
+ "sphinx-copybutton",
+ "numpydoc",
+ "sphinx-prompt",
+ "plotly",
+ "polars",
+ "pooch",
+ "sphinxext-opengraph",
+ "sphinx-remove-toctrees",
+ "sphinx-design",
+ "pydata-sphinx-theme",
+ ],
+ "pip_dependencies": [
+ "jupyterlite-sphinx",
+ "jupyterlite-pyodide-kernel",
+ "sphinxcontrib-sass",
+ ],
+ "package_constraints": {
+ "python": "3.9",
+ },
+ },
+ {
+ "name": "pymin_conda_forge",
+ "type": "conda",
+ "tag": "arm",
+ "folder": "build_tools/cirrus",
+ "platform": "linux-aarch64",
+ "channels": ["conda-forge"],
+ "conda_dependencies": remove_from(
+ common_dependencies_without_coverage, ["pandas", "pyamg"]
+ )
+ + ["pip", "ccache"],
+ "package_constraints": {
+ "python": "3.9",
+ },
+ },
+ {
+ "name": "debian_atlas_32bit",
+ "type": "pip",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "pip_dependencies": [
+ "cython",
+ "joblib",
+ "threadpoolctl",
+ "pytest",
+ "pytest-cov",
+ "ninja",
+ "meson-python",
+ ],
+ "package_constraints": {
+ "joblib": "min",
+ "threadpoolctl": "3.1.0",
+ "pytest": "min",
+ "pytest-cov": "min",
+ # no pytest-xdist because it causes issue on 32bit
+ "cython": "min",
+ },
+ # same Python version as in debian-32 build
+ "python_version": "3.11.2",
+ },
+ {
+ "name": "ubuntu_atlas",
+ "type": "pip",
+ "tag": "main-ci",
+ "folder": "build_tools/azure",
+ "pip_dependencies": [
+ "cython",
+ "joblib",
+ "threadpoolctl",
+ "pytest",
+ "pytest-xdist",
+ "ninja",
+ "meson-python",
+ ],
+ "package_constraints": {
+ "joblib": "min",
+ "threadpoolctl": "min",
+ "cython": "min",
+ },
+ "python_version": "3.10.4",
+ },
+]
+
+
+def execute_command(command_list):
+ logger.debug(" ".join(command_list))
+ proc = subprocess.Popen(
+ command_list, stdout=subprocess.PIPE, stderr=subprocess.PIPE
+ )
+
+ out, err = proc.communicate()
+ out, err = out.decode(errors="replace"), err.decode(errors="replace")
+
+ if proc.returncode != 0:
+ command_str = " ".join(command_list)
+ raise RuntimeError(
+ "Command exited with non-zero exit code.\n"
+ "Exit code: {}\n"
+ "Command:\n{}\n"
+ "stdout:\n{}\n"
+ "stderr:\n{}\n".format(proc.returncode, command_str, out, err)
+ )
+ logger.log(TRACE, out)
+ return out
+
+
+def get_package_with_constraint(package_name, build_metadata, uses_pip=False):
+ build_package_constraints = build_metadata.get("package_constraints")
+ if build_package_constraints is None:
+ constraint = None
+ else:
+ constraint = build_package_constraints.get(package_name)
+
+ constraint = constraint or default_package_constraints.get(package_name)
+
+ if constraint is None:
+ return package_name
+
+ comment = ""
+ if constraint == "min":
+ constraint = execute_command(
+ [sys.executable, "sklearn/_min_dependencies.py", package_name]
+ ).strip()
+ comment = " # min"
+
+ if re.match(r"\d[.\d]*", constraint):
+ equality = "==" if uses_pip else "="
+ constraint = equality + constraint
+
+ return f"{package_name}{constraint}{comment}"
+
+
+environment = Environment(trim_blocks=True, lstrip_blocks=True)
+environment.filters["get_package_with_constraint"] = get_package_with_constraint
+
+
+def get_conda_environment_content(build_metadata):
+ template = environment.from_string(
+ """
+# DO NOT EDIT: this file is generated from the specification found in the
+# following script to centralize the configuration for CI builds:
+# build_tools/update_environments_and_lock_files.py
+channels:
+ {% for channel in build_metadata['channels'] %}
+ - {{ channel }}
+ {% endfor %}
+dependencies:
+ {% for conda_dep in build_metadata['conda_dependencies'] %}
+ - {{ conda_dep | get_package_with_constraint(build_metadata) }}
+ {% endfor %}
+ {% if build_metadata['pip_dependencies'] %}
+ - pip
+ - pip:
+ {% for pip_dep in build_metadata.get('pip_dependencies', []) %}
+ - {{ pip_dep | get_package_with_constraint(build_metadata, uses_pip=True) }}
+ {% endfor %}
+ {% endif %}""".strip()
+ )
+ return template.render(build_metadata=build_metadata)
+
+
+def write_conda_environment(build_metadata):
+ content = get_conda_environment_content(build_metadata)
+ build_name = build_metadata["name"]
+ folder_path = Path(build_metadata["folder"])
+ output_path = folder_path / f"{build_name}_environment.yml"
+ logger.debug(output_path)
+ output_path.write_text(content)
+
+
+def write_all_conda_environments(build_metadata_list):
+ for build_metadata in build_metadata_list:
+ write_conda_environment(build_metadata)
+
+
+def conda_lock(environment_path, lock_file_path, platform):
+ execute_command(
+ [
+ "conda-lock",
+ "lock",
+ "--mamba",
+ "--kind",
+ "explicit",
+ "--platform",
+ platform,
+ "--file",
+ str(environment_path),
+ "--filename-template",
+ str(lock_file_path),
+ ]
+ )
+
+
+def create_conda_lock_file(build_metadata):
+ build_name = build_metadata["name"]
+ folder_path = Path(build_metadata["folder"])
+ environment_path = folder_path / f"{build_name}_environment.yml"
+ platform = build_metadata["platform"]
+ lock_file_basename = build_name
+ if not lock_file_basename.endswith(platform):
+ lock_file_basename = f"{lock_file_basename}_{platform}"
+
+ lock_file_path = folder_path / f"{lock_file_basename}_conda.lock"
+ conda_lock(environment_path, lock_file_path, platform)
+
+
+def write_all_conda_lock_files(build_metadata_list):
+ for build_metadata in build_metadata_list:
+ logger.info(f"# Locking dependencies for {build_metadata['name']}")
+ create_conda_lock_file(build_metadata)
+
+
+def get_pip_requirements_content(build_metadata):
+ template = environment.from_string(
+ """
+# DO NOT EDIT: this file is generated from the specification found in the
+# following script to centralize the configuration for CI builds:
+# build_tools/update_environments_and_lock_files.py
+{% for pip_dep in build_metadata['pip_dependencies'] %}
+{{ pip_dep | get_package_with_constraint(build_metadata, uses_pip=True) }}
+{% endfor %}""".strip()
+ )
+ return template.render(build_metadata=build_metadata)
+
+
+def write_pip_requirements(build_metadata):
+ build_name = build_metadata["name"]
+ content = get_pip_requirements_content(build_metadata)
+ folder_path = Path(build_metadata["folder"])
+ output_path = folder_path / f"{build_name}_requirements.txt"
+ logger.debug(output_path)
+ output_path.write_text(content)
+
+
+def write_all_pip_requirements(build_metadata_list):
+ for build_metadata in build_metadata_list:
+ write_pip_requirements(build_metadata)
+
+
+def pip_compile(pip_compile_path, requirements_path, lock_file_path):
+ execute_command(
+ [
+ str(pip_compile_path),
+ "--upgrade",
+ str(requirements_path),
+ "-o",
+ str(lock_file_path),
+ ]
+ )
+
+
+def write_pip_lock_file(build_metadata):
+ build_name = build_metadata["name"]
+ python_version = build_metadata["python_version"]
+ environment_name = f"pip-tools-python{python_version}"
+ # To make sure that the Python used to create the pip lock file is the same
+ # as the one used during the CI build where the lock file is used, we first
+ # create a conda environment with the correct Python version and
+ # pip-compile and run pip-compile in this environment
+
+ execute_command(
+ [
+ "conda",
+ "create",
+ "-c",
+ "conda-forge",
+ "-n",
+ f"pip-tools-python{python_version}",
+ f"python={python_version}",
+ "pip-tools",
+ "-y",
+ ]
+ )
+
+ json_output = execute_command(["conda", "info", "--json"])
+ conda_info = json.loads(json_output)
+ environment_folder = [
+ each for each in conda_info["envs"] if each.endswith(environment_name)
+ ][0]
+ environment_path = Path(environment_folder)
+ pip_compile_path = environment_path / "bin" / "pip-compile"
+
+ folder_path = Path(build_metadata["folder"])
+ requirement_path = folder_path / f"{build_name}_requirements.txt"
+ lock_file_path = folder_path / f"{build_name}_lock.txt"
+ pip_compile(pip_compile_path, requirement_path, lock_file_path)
+
+
+def write_all_pip_lock_files(build_metadata_list):
+ for build_metadata in build_metadata_list:
+ logger.info(f"# Locking dependencies for {build_metadata['name']}")
+ write_pip_lock_file(build_metadata)
+
+
+def check_conda_lock_version():
+ # Check that the installed conda-lock version is consistent with _min_dependencies.
+ expected_conda_lock_version = execute_command(
+ [sys.executable, "sklearn/_min_dependencies.py", "conda-lock"]
+ ).strip()
+
+ installed_conda_lock_version = version("conda-lock")
+ if installed_conda_lock_version != expected_conda_lock_version:
+ raise RuntimeError(
+ f"Expected conda-lock version: {expected_conda_lock_version}, got:"
+ f" {installed_conda_lock_version}"
+ )
+
+
+def check_conda_version():
+ # Avoid issues with glibc (https://github.com/conda/conda-lock/issues/292)
+ # or osx (https://github.com/conda/conda-lock/issues/408) virtual package.
+ # The glibc one has been fixed in conda 23.1.0 and the osx has been fixed
+ # in conda 23.7.0.
+ conda_info_output = execute_command(["conda", "info", "--json"])
+
+ conda_info = json.loads(conda_info_output)
+ conda_version = Version(conda_info["conda_version"])
+
+ if Version("22.9.0") < conda_version < Version("23.7"):
+ raise RuntimeError(
+ f"conda version should be <= 22.9.0 or >= 23.7 got: {conda_version}"
+ )
+
+
+@click.command()
+@click.option(
+ "--select-build",
+ default="",
+ help=(
+ "Regex to filter the builds we want to update environment and lock files. By"
+ " default all the builds are selected."
+ ),
+)
+@click.option(
+ "--skip-build",
+ default=None,
+ help="Regex to skip some builds from the builds selected by --select-build",
+)
+@click.option(
+ "--select-tag",
+ default=None,
+ help=(
+ "Tag to filter the builds, e.g. 'main-ci' or 'scipy-dev'. "
+ "This is an additional filtering on top of --select-build."
+ ),
+)
+@click.option(
+ "-v",
+ "--verbose",
+ is_flag=True,
+ help="Print commands executed by the script",
+)
+@click.option(
+ "-vv",
+ "--very-verbose",
+ is_flag=True,
+ help="Print output of commands executed by the script",
+)
+def main(select_build, skip_build, select_tag, verbose, very_verbose):
+ if verbose:
+ logger.setLevel(logging.DEBUG)
+ if very_verbose:
+ logger.setLevel(TRACE)
+ handler.setLevel(TRACE)
+ check_conda_lock_version()
+ check_conda_version()
+
+ filtered_build_metadata_list = [
+ each for each in build_metadata_list if re.search(select_build, each["name"])
+ ]
+ if select_tag is not None:
+ filtered_build_metadata_list = [
+ each for each in build_metadata_list if each["tag"] == select_tag
+ ]
+ if skip_build is not None:
+ filtered_build_metadata_list = [
+ each
+ for each in filtered_build_metadata_list
+ if not re.search(skip_build, each["name"])
+ ]
+
+ selected_build_info = "\n".join(
+ f" - {each['name']}, type: {each['type']}, tag: {each['tag']}"
+ for each in filtered_build_metadata_list
+ )
+ selected_build_message = (
+ f"# {len(filtered_build_metadata_list)} selected builds\n{selected_build_info}"
+ )
+ logger.info(selected_build_message)
+
+ filtered_conda_build_metadata_list = [
+ each for each in filtered_build_metadata_list if each["type"] == "conda"
+ ]
+
+ if filtered_conda_build_metadata_list:
+ logger.info("# Writing conda environments")
+ write_all_conda_environments(filtered_conda_build_metadata_list)
+ logger.info("# Writing conda lock files")
+ write_all_conda_lock_files(filtered_conda_build_metadata_list)
+
+ filtered_pip_build_metadata_list = [
+ each for each in filtered_build_metadata_list if each["type"] == "pip"
+ ]
+ if filtered_pip_build_metadata_list:
+ logger.info("# Writing pip requirements")
+ write_all_pip_requirements(filtered_pip_build_metadata_list)
+ logger.info("# Writing pip lock files")
+ write_all_pip_lock_files(filtered_pip_build_metadata_list)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/auto_building_tools/build_tools/wheels/build_wheels.sh b/auto_building_tools/build_tools/wheels/build_wheels.sh
new file mode 100644
index 0000000..f2ed849
--- /dev/null
+++ b/auto_building_tools/build_tools/wheels/build_wheels.sh
@@ -0,0 +1,64 @@
+#!/bin/bash
+
+set -e
+set -x
+
+# Set environment variables to make our wheel build easier to reproduce byte
+# for byte from source. See https://reproducible-builds.org/. The long term
+# motivation would be to be able to detect supply chain attacks.
+#
+# In particular we set SOURCE_DATE_EPOCH to the commit date of the last commit.
+#
+# XXX: setting those environment variables is not enough. See the following
+# issue for more details on what remains to do:
+# https://github.com/scikit-learn/scikit-learn/issues/28151
+export SOURCE_DATE_EPOCH=$(git log -1 --pretty=%ct)
+export PYTHONHASHSEED=0
+
+# OpenMP is not present on macOS by default
+if [[ $(uname) == "Darwin" ]]; then
+ # Make sure to use a libomp version binary compatible with the oldest
+ # supported version of the macos SDK as libomp will be vendored into the
+ # scikit-learn wheels for macos.
+
+ if [[ "$CIBW_BUILD" == *-macosx_arm64 ]]; then
+ if [[ $(uname -m) == "x86_64" ]]; then
+ # arm64 builds must cross compile because the CI instance is x86
+ # This turns off the computation of the test program in
+ # sklearn/_build_utils/pre_build_helpers.py
+ export PYTHON_CROSSENV=1
+ fi
+ # SciPy requires 12.0 on arm to prevent kernel panics
+ # https://github.com/scipy/scipy/issues/14688
+ # We use the same deployment target to match SciPy.
+ export MACOSX_DEPLOYMENT_TARGET=12.0
+ OPENMP_URL="https://anaconda.org/conda-forge/llvm-openmp/11.1.0/download/osx-arm64/llvm-openmp-11.1.0-hf3c4609_1.tar.bz2"
+ else
+ export MACOSX_DEPLOYMENT_TARGET=10.9
+ OPENMP_URL="https://anaconda.org/conda-forge/llvm-openmp/11.1.0/download/osx-64/llvm-openmp-11.1.0-hda6cdc1_1.tar.bz2"
+ fi
+
+ sudo conda create -n build $OPENMP_URL
+ PREFIX="$CONDA_HOME/envs/build"
+
+ export CC=/usr/bin/clang
+ export CXX=/usr/bin/clang++
+ export CPPFLAGS="$CPPFLAGS -Xpreprocessor -fopenmp"
+ export CFLAGS="$CFLAGS -I$PREFIX/include"
+ export CXXFLAGS="$CXXFLAGS -I$PREFIX/include"
+ export LDFLAGS="$LDFLAGS -Wl,-rpath,$PREFIX/lib -L$PREFIX/lib -lomp"
+fi
+
+if [[ "$CIBW_FREE_THREADED_SUPPORT" =~ [tT]rue ]]; then
+ # Numpy, scipy, Cython only have free-threaded wheels on scientific-python-nightly-wheels
+ # TODO: remove this after CPython 3.13 is released (scheduled October 2024)
+ # and our dependencies have free-threaded wheels on PyPI
+ export CIBW_BUILD_FRONTEND='pip; args: --pre --extra-index-url "https://pypi.anaconda.org/scientific-python-nightly-wheels/simple" --only-binary :all:'
+fi
+
+# The version of the built dependencies are specified
+# in the pyproject.toml file, while the tests are run
+# against the most recent version of the dependencies
+
+python -m pip install cibuildwheel
+python -m cibuildwheel --output-dir wheelhouse
diff --git a/auto_building_tools/build_tools/wheels/cibw_before_test.sh b/auto_building_tools/build_tools/wheels/cibw_before_test.sh
new file mode 100644
index 0000000..b888eac
--- /dev/null
+++ b/auto_building_tools/build_tools/wheels/cibw_before_test.sh
@@ -0,0 +1,10 @@
+#!/bin/bash
+
+set -e
+set -x
+
+FREE_THREADED_BUILD="$(python -c"import sysconfig; print(bool(sysconfig.get_config_var('Py_GIL_DISABLED')))")"
+if [[ $FREE_THREADED_BUILD == "True" ]]; then
+ # TODO: remove when numpy, scipy and pandas have releases with free-threaded wheels
+ python -m pip install --pre --extra-index https://pypi.anaconda.org/scientific-python-nightly-wheels/simple numpy scipy pandas --only-binary :all:
+fi
diff --git a/auto_building_tools/build_tools/wheels/test_wheels.sh b/auto_building_tools/build_tools/wheels/test_wheels.sh
new file mode 100644
index 0000000..da2c458
--- /dev/null
+++ b/auto_building_tools/build_tools/wheels/test_wheels.sh
@@ -0,0 +1,26 @@
+#!/bin/bash
+
+set -e
+set -x
+
+python -c "import joblib; print(f'Number of cores (physical): \
+{joblib.cpu_count()} ({joblib.cpu_count(only_physical_cores=True)})')"
+
+FREE_THREADED_BUILD="$(python -c"import sysconfig; print(bool(sysconfig.get_config_var('Py_GIL_DISABLED')))")"
+if [[ $FREE_THREADED_BUILD == "True" ]]; then
+ # TODO: delete when importing numpy no longer enables the GIL
+ # setting to zero ensures the GIL is disabled while running the
+ # tests under free-threaded python
+ export PYTHON_GIL=0
+fi
+
+# Test that there are no links to system libraries in the
+# threadpoolctl output section of the show_versions output:
+python -c "import sklearn; sklearn.show_versions()"
+
+if pip show -qq pytest-xdist; then
+ XDIST_WORKERS=$(python -c "import joblib; print(joblib.cpu_count(only_physical_cores=True))")
+ pytest --pyargs sklearn -n $XDIST_WORKERS
+else
+ pytest --pyargs sklearn
+fi
diff --git a/auto_building_tools/citation/CITATION.bib.template b/auto_building_tools/citation/CITATION.bib.template
new file mode 100644
index 0000000..a8a4734
--- /dev/null
+++ b/auto_building_tools/citation/CITATION.bib.template
@@ -0,0 +1,82 @@
+{#-
+# https://docs.github.com/en/repositories/managing-your-repositorys-settings-and-features/customizing-your-repository/about-citation-files
+-#}
+{#-
+# Authors: The scikit-plots developers
+# SPDX-License-Identifier: BSD-3-Clause
+-#}
+% ---------------------------------------------------------
+% CITATION.bib file for {{ library_name }}
+% This file provides citation information for users
+% who want to cite the library, related papers, and books.
+% ---------------------------------------------------------
+{% if include_misc %}
+@misc{{ '{' }}{{ library_name }}:v{{ version }},
+ {#- The author(s) or maintainer(s) of the Python library #}
+ author = { {{ author }} },
+ {#- The title of your Python library (name it appropriately) #}
+ title = { {{ library_title }} },
+ {#- Year of the library release or the latest significant update #}
+ year = { {{ release_year }} },
+ {#- Optional: Add the version number of your library #}
+ version = { {{ version }} },
+ {#- The URL to your project repository (GitHub, GitLab, etc.) #}
+ url = { {{ url }} },
+ {#- You can provide a short note, such as the purpose of the library or a brief description #}
+ note = { {{ note }} },
+ {#- Optional: Add the DOI if your project has one (e.g., if you've archived it in Zenodo) #}
+ doi = { {{ doi }} }
+}
+{%- endif %}
+
+{#-
+% ---------------------------------------------------------
+% ARTICLE TEMPLATE (Future use):
+% Commented out for now, uncomment when you need it.
+% ---------------------------------------------------------
+#}
+{%- if include_article %}
+@article{{ '{' }}{{ article_key }}:{{ article_year }},
+ author = { {{ article_authors }} },
+ title = { {{ article_title }} },
+ journal = { {{ article_journal }} },
+ year = { {{ article_year }} },
+ volume = { {{ article_volume }} },
+ number = { {{ article_number }} },
+ pages = { {{ article_pages }} },
+ doi = { {{ article_doi }} },
+ url = { {{ article_url }} }
+}
+{%- endif %}
+
+{#-
+% ---------------------------------------------------------
+% BOOK TEMPLATE (Future use):
+% Commented out for now, uncomment when you need it.
+% ---------------------------------------------------------
+#}
+{%- if include_book %}
+@book{{ '{' }}{{ book_key }}:{{ book_year }},
+ author = { {{ book_author }} },
+ title = { {{ book_title }} },
+ publisher = { {{ book_publisher }} },
+ year = { {{ book_year }} },
+ edition = { {{ book_edition }} },
+ url = { {{ book_url }} },
+ isbn = { {{ book_isbn }} }
+}
+{%- endif %}
+
+{#-
+% ---------------------------------------------------------
+% GUIDELINES FOR FUTURE UPDATES
+% ---------------------------------------------------------
+% 1. "author" field: List all the authors or contributors of the work.
+% Format: {Last Name, First Name and Last Name, First Name}.
+% 2. "title" field: For articles, use the title of the paper. For books, use the book title.
+% 3. "year" field: Update the year accordingly.
+% 4. "journal" and "publisher": For articles, include the journal name. For books, include the publisher.
+% 5. "volume", "number", "pages": Applicable only to journal articles.
+% 6. "doi" and "url": Always include DOI if available; otherwise, provide a URL.
+% 7. "isbn": If your book has an ISBN, include it to make the citation more complete.
+-#}
\ No newline at end of file
diff --git a/auto_building_tools/citation/CITATION.cff.template b/auto_building_tools/citation/CITATION.cff.template
new file mode 100644
index 0000000..8433fe6
--- /dev/null
+++ b/auto_building_tools/citation/CITATION.cff.template
@@ -0,0 +1,85 @@
+{#-
+# https://docs.github.com/en/repositories/managing-your-repositorys-settings-and-features/customizing-your-repository/about-citation-files
+-#}
+{#-
+# Authors: The scikit-plots developers
+# SPDX-License-Identifier: BSD-3-Clause
+-#}
+# ---------------------------------------------------------
+# CITATION.cff file for {{ library_name }}
+# This file provides citation information for users
+# who want to cite the library, related papers, and books.
+# ---------------------------------------------------------
+
+cff-version: 1.2.0 # The version of the CFF format used.
+message: "If you use this software, please cite it using the following metadata." {# Optional message to users. #}
+title: "{{ library_title | trim }}" {# The name of your software. #}
+version: "{{ version | trim }}" {# The current version of your software. #}
+doi: "{{ doi | trim }}" {# Optional DOI (if archived on platforms like Zenodo). #}
+date-released: "{{ release_date | trim }}" {# Date when this version was released. #}
+
+# Authors and contributors of the software project.
+authors:
+ - team: "{{ author | trim }}"
+ website: "{{ url | trim }}"
+ {#- given-names: "{{ author_given | trim }}" -#}
+ {#- Optional, but recommended. -#}
+ {#- affiliation: "{{ author_affiliation | trim }}" -#}
+ {#- Optional, if you have an ORCID. -#}
+ {#- orcid: "{{ author_orcid | trim }}" -#}
+
+ {#- Uncomment this section for additional contributors when needed. -#}
+ {#-
+ - family-names: ContributorLastName
+ given-names: ContributorFirstName
+ affiliation: Contributor Organization
+ orcid: https://orcid.org/0000-0002-9876-5432 #}
+
+# Repository information
+{#- The URL to your software repository (GitHub, GitLab, etc.). #}
+repository-code: "{{ repository_url | trim }}"
+{#- Optional: Link to archived artifacts (e.g., Zenodo DOI). #}
+repository-artifact: "{{ artifact_url | trim }}"
+
+# License for your software
+{#- The license under which your software is released. -#}
+license: "{{ license | trim }}"
+type: software
+url: "{{ url | trim }}"
+
+# Keywords related to your software
+keywords:
+ {#- This allows a list of keywords. #}
+ - {{ keywords | join("\n - ") | trim }}
+
+{#-
+# Optionally, add references to any related articles or papers
+# Uncomment and fill in the details when needed.
+# references:
+# - type: article
+# authors:
+# - family-names: AuthorLastName
+# given-names: AuthorFirstName
+# title: "Title of Related Research Paper"
+# year: 2024
+# date-published: 2007-06-18
+# journal: "Journal Name"
+# volume: x
+# issue: x
+# start: xx
+# end: xx
+# doi: 10.1016/j.dat.xxxx.xx.xxx
+# publisher:
+# name: IEEE Computer Society
+# website: ''
+# abstract: ''
+#
+# - type: book
+# authors:
+# - family-names: AuthorLastName
+# given-names: AuthorFirstName
+# title: "Title of Related Book"
+# publisher: "Publisher Name"
+# year: 2024
+# isbn: "978-1-234-56789-0"
+-#}
\ No newline at end of file
diff --git a/auto_building_tools/citation/citation_ref.py b/auto_building_tools/citation/citation_ref.py
new file mode 100644
index 0000000..81576d7
--- /dev/null
+++ b/auto_building_tools/citation/citation_ref.py
@@ -0,0 +1,79 @@
+import os
+from datetime import datetime
+from jinja2 import Environment, FileSystemLoader
+
+current_dir = os.path.dirname(__file__)
+output_path = os.path.join(current_dir, '../../')
+os.makedirs(output_path, exist_ok=True)
+
+# Get the current date and time
+now = datetime.now()
+formatted_date = now.strftime("%Y-%m-%d") # Outputs: '2024-10-27'
+# year_str = str(now.year)
+# month_str = str(now.month).zfill(2) # Ensures month has 2 digits (e.g., '01')
+# day_str = str(now.day).zfill(2)
+
+# Step 1: Load your template
+env = Environment(loader=FileSystemLoader(current_dir))
+template_bib = env.get_template('CITATION.bib.template')
+template_cff = env.get_template('CITATION.cff.template')
+
+# Step 2: Define the values to render the template
+context = {
+ # misc
+ "include_misc": True,
+ "library_name": "scikit-plots",
+ "author": "scikit-plots developers",
+ "library_title": "scikit-plots: Machine Learning Visualization in Python",
+ "release_year": "2024",
+ "version": "latest",
+ "url": "https://scikit-plots.github.io",
+ "note": "Scikit-plot is the result of an unartistic data scientist's dreadful realization that visualization is one of the most crucial components in the data science process, not just a mere afterthought.",
+ "doi": "10.5281/zenodo.13367000",
+ # article
+ "include_article": False,
+ "article_key": "yourarticle2024",
+ "article_authors": "Author One and Author Two",
+ "article_title": "Title of the Research Paper Associated with the Library",
+ "article_journal": "Journal of Research",
+ "article_year": "2024",
+ "article_volume": "10",
+ "article_number": "2",
+ "article_pages": "123-134",
+ "article_doi": "10.1234/journal.doi",
+ "article_url": "https://doi.org/10.1234/journal.doi",
+ # book
+ "include_book": False,
+ "book_key": "yourbook2024",
+ "book_author": "Author Name",
+ "book_title": "The Python Library Name: Comprehensive Guide",
+ "book_publisher": "Example Publishing House",
+ "book_year": "2024",
+ "book_edition": "2nd",
+ "book_url": "https://linktothebook.com",
+ "book_isbn": "978-3-16-148410-0",
+ # cff
+ "library_title": "scikit-plots: Machine Learning Visualization in Python",
+ "release_date": formatted_date,
+ "author_family": "Çelik",
+ "author_given": "Muhammed",
+ "author_affiliation": "scikit-plots",
+ "author_orcid": "https://orcid.org/0000-0001-2345-6789",
+ "repository_url": "https://github.com/scikit-plots/scikit-plots",
+ "artifact_url": "https://zenodo.org/records/13367000",
+ "license": "BSD-3-Clause",
+ "keywords": ["Software", "Python", "scikit-plots", "Machine Learning Visualization"]
+}
+
+for template, output_fname in zip([template_bib, template_cff], ['CITATION.bib', 'CITATION.cff']):
+ # Step 3: Render the template with actual values
+ output = template.render(context)
+
+ # Step 4: Remove extra newlines
+ # output = "\n".join([line for line in output.splitlines() if line.strip()])
+
+ # Step 5: Print the rendered result or save it to a file
+ with open(os.path.join(output_path, output_fname), 'w') as output_file:
+ output_file.write(output)
+
+ print(f"{os.path.join(output_path, output_fname)} file created successfully.")
\ No newline at end of file
diff --git a/auto_building_tools/maint_tools/sort_whats_new.py b/auto_building_tools/maint_tools/sort_whats_new.py
new file mode 100644
index 0000000..7241059
--- /dev/null
+++ b/auto_building_tools/maint_tools/sort_whats_new.py
@@ -0,0 +1,43 @@
+#!/usr/bin/env python
+# Sorts what's new entries with per-module headings.
+# Pass what's new entries on stdin.
+
+import re
+import sys
+from collections import defaultdict
+
+LABEL_ORDER = ["MajorFeature", "Feature", "Efficiency", "Enhancement", "Fix", "API"]
+
+
+def entry_sort_key(s):
+ if s.startswith("- |"):
+ return LABEL_ORDER.index(s.split("|")[1])
+ else:
+ return -1
+
+
+# discard headings and other non-entry lines
+text = "".join(l for l in sys.stdin if l.startswith("- ") or l.startswith(" "))
+
+bucketed = defaultdict(list)
+
+for entry in re.split("\n(?=- )", text.strip()):
+ modules = re.findall(
+ r":(?:func|meth|mod|class):" r"`(?:[^<`]*<|~)?(?:sklearn.)?([a-z]\w+)", entry
+ )
+ modules = set(modules)
+ if len(modules) > 1:
+ key = "Multiple modules"
+ elif modules:
+ key = ":mod:`sklearn.%s`" % next(iter(modules))
+ else:
+ key = "Miscellaneous"
+ bucketed[key].append(entry)
+ entry = entry.strip() + "\n"
+
+everything = []
+for key, bucket in sorted(bucketed.items()):
+ everything.append(key + "\n" + "." * len(key))
+ bucket.sort(key=entry_sort_key)
+ everything.extend(bucket)
+print("\n\n".join(everything))
diff --git a/auto_building_tools/maint_tools/update_tracking_issue.py b/auto_building_tools/maint_tools/update_tracking_issue.py
new file mode 100644
index 0000000..b40e822
--- /dev/null
+++ b/auto_building_tools/maint_tools/update_tracking_issue.py
@@ -0,0 +1,163 @@
+"""Creates or updates an issue if the CI fails. This is useful to keep track of
+scheduled jobs that are failing repeatedly.
+
+This script depends on:
+- `defusedxml` for safer parsing for xml
+- `PyGithub` for interacting with GitHub
+
+The GitHub token only requires the `repo:public_repo` scope are described in
+https://docs.github.com/en/developers/apps/building-oauth-apps/scopes-for-oauth-apps#available-scopes.
+This scope allows the bot to create and edit its own issues. It is best to use a
+github account that does **not** have commit access to the public repo.
+"""
+
+import argparse
+import sys
+from datetime import datetime, timezone
+from pathlib import Path
+
+import defusedxml.ElementTree as ET
+from github import Github
+
+parser = argparse.ArgumentParser(
+ description="Create or update issue from JUnit test results from pytest"
+)
+parser.add_argument(
+ "bot_github_token", help="Github token for creating or updating an issue"
+)
+parser.add_argument("ci_name", help="Name of CI run instance")
+parser.add_argument("issue_repo", help="Repo to track issues")
+parser.add_argument("link_to_ci_run", help="URL to link to")
+parser.add_argument("--junit-file", help="JUnit file to determine if tests passed")
+parser.add_argument(
+ "--tests-passed",
+ help=(
+ "If --tests-passed is true, then the original issue is closed if the issue "
+ "exists. If tests-passed is false, then the an issue is updated or created."
+ ),
+)
+parser.add_argument(
+ "--auto-close",
+ help=(
+ "If --auto-close is false, then issues will not auto close even if the tests"
+ " pass."
+ ),
+ default="true",
+)
+
+args = parser.parse_args()
+
+if args.junit_file is not None and args.tests_passed is not None:
+ print("--junit-file and --test-passed can not be set together")
+ sys.exit(1)
+
+if args.junit_file is None and args.tests_passed is None:
+ print("Either --junit-file or --test-passed must be passed in")
+ sys.exit(1)
+
+gh = Github(args.bot_github_token)
+issue_repo = gh.get_repo(args.issue_repo)
+dt_now = datetime.now(tz=timezone.utc)
+date_str = dt_now.strftime("%b %d, %Y")
+title_query = f"CI failed on {args.ci_name}"
+title = f"⚠️ {title_query} (last failure: {date_str}) ⚠️"
+
+
+def get_issue():
+ login = gh.get_user().login
+ issues = gh.search_issues(
+ f"repo:{args.issue_repo} {title_query} in:title state:open author:{login}"
+ )
+ first_page = issues.get_page(0)
+ # Return issue if it exist
+ return first_page[0] if first_page else None
+
+
+def create_or_update_issue(body=""):
+ # Interact with GitHub API to create issue
+ link = f"[{args.ci_name}]({args.link_to_ci_run})"
+ issue = get_issue()
+
+ max_body_length = 60_000
+ original_body_length = len(body)
+ # Avoid "body is too long (maximum is 65536 characters)" error from github REST API
+ if original_body_length > max_body_length:
+ body = (
+ f"{body[:max_body_length]}\n...\n"
+ f"Body was too long ({original_body_length} characters) and was shortened"
+ )
+
+ if issue is None:
+ # Create new issue
+ header = f"**CI failed on {link}** ({date_str})"
+ issue = issue_repo.create_issue(title=title, body=f"{header}\n{body}")
+ print(f"Created issue in {args.issue_repo}#{issue.number}")
+ sys.exit()
+ else:
+ # Update existing issue
+ header = f"**CI is still failing on {link}** ({date_str})"
+ issue.edit(title=title, body=f"{header}\n{body}")
+ print(f"Commented on issue: {args.issue_repo}#{issue.number}")
+ sys.exit()
+
+
+def close_issue_if_opened():
+ print("Test has no failures!")
+ issue = get_issue()
+ if issue is not None:
+ header_str = "## CI is no longer failing!"
+ comment_str = (
+ f"{header_str} ✅\n\n[Successful run]({args.link_to_ci_run}) on {date_str}"
+ )
+
+ print(f"Commented on issue #{issue.number}")
+ # New comment if "## CI is no longer failing!" comment does not exist
+ # If it does exist update the original comment which includes the new date
+ for comment in issue.get_comments():
+ if comment.body.startswith(header_str):
+ comment.edit(body=comment_str)
+ break
+ else: # no break
+ issue.create_comment(body=comment_str)
+
+ if args.auto_close.lower() == "true":
+ print(f"Closing issue #{issue.number}")
+ issue.edit(state="closed")
+ sys.exit()
+
+
+if args.tests_passed is not None:
+ if args.tests_passed.lower() == "true":
+ close_issue_if_opened()
+ else:
+ create_or_update_issue()
+
+junit_path = Path(args.junit_file)
+if not junit_path.exists():
+ body = "Unable to find junit file. Please see link for details."
+ create_or_update_issue(body)
+
+# Find failures in junit file
+tree = ET.parse(args.junit_file)
+failure_cases = []
+
+# Check if test collection failed
+error = tree.find("./testsuite/testcase/error")
+if error is not None:
+ # Get information for test collection error
+ failure_cases.append("Test Collection Failure")
+
+for item in tree.iter("testcase"):
+ failure = item.find("failure")
+ if failure is None:
+ continue
+
+ failure_cases.append(item.attrib["name"])
+
+if not failure_cases:
+ close_issue_if_opened()
+
+# Create content for issue
+body_list = [f"- {case}" for case in failure_cases]
+body = "\n".join(body_list)
+create_or_update_issue(body)
diff --git a/auto_building_tools/maint_tools/whats_missing.sh b/auto_building_tools/maint_tools/whats_missing.sh
new file mode 100644
index 0000000..6627778
--- /dev/null
+++ b/auto_building_tools/maint_tools/whats_missing.sh
@@ -0,0 +1,61 @@
+#!/bin/bash
+# This script helps identify pull requests that were merged without a what's
+# new entry, where one would be appropriate.
+
+if [ $# -ne 2 ]
+then
+ echo "Usage: GITHUB_TOKEN=... $0